Molecular characteristics of low-grade serous carcinoma in effusions
Abstract
Objective
The molecular characteristics of low-grade serous carcinoma (LGSC) in serous effusions have not been studied previously. The present study analysed the molecular profile of LGSC at this anatomical site.
Methods
Specimens consisted of a series of 17 serous effusions (15 peritoneal, 2 pleural) from 16 patients, of which 15 were LGSC and 2 serous borderline tumour (SBT) who later progressed to LGSC. For comparative purposes, 9 surgical specimens from 6 patients with LGSC were analysed. Fresh-frozen cell pellets and surgical specimens underwent targeted next-generation sequencing covering 50 unique genes.
Results
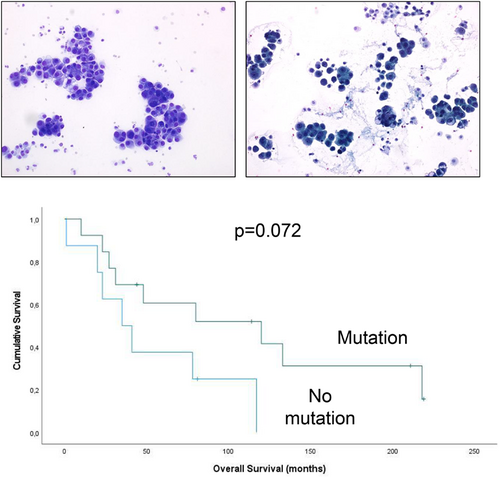
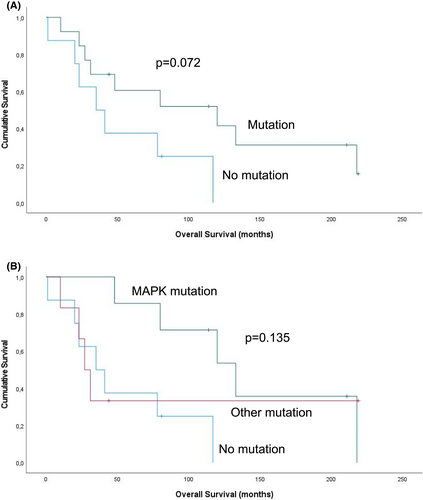
Mutations were found in tumours from 14 of the 22 patients, of whom 4 had 2 different mutations and 10 had a single mutation. Overall, the most common mutations were in KRAS (n = 3) and BRAF (n = 3), followed by NRAS (n = 2), CDK2NA (n = 2), TP53 (n = 2), ATM (n = 2). Mutations in MET, STK11, ERBB2 and FLT3 were found in one case each. Patient-matched specimens had the same molecular profile. Both effusions with TP53 mutation had concomitant ATM mutation, and both stained immunohistochemically with a wild-type pattern. The absence of mutations was associated with a trend for shorter overall survival in univariate analysis (p = 0.072).
Conclusions
The molecular alterations in LGSCs in serous effusions are consistent with those found in solid tumours, with frequent alterations in the mitogen-activated protein kinase pathway. Mutations in LGSC may be a marker of better outcomes.
Graphical Abstract
1 INTRODUCTION
Globally, ovarian cancer is ranked as the eighth most common and the eighth most lethal cancer in women. In 2020, 313,959 new diagnoses and 207,252 deaths from this disease were reported, corresponding to 3.4% of cancer diagnoses and 4.7% of cancer-related deaths in females.1 The majority (> 90%) of tubo-ovarian cancers are carcinomas, of which the main histotypes are high-grade serous carcinoma (HGSC), low-grade serous carcinoma (LGSC), clear cell carcinoma (CCC), endometrioid carcinoma (EC) and mucinous carcinoma (MC). HGSC accounts for 70% of cases.2
Low-grade serous carcinoma represents an uncommon histological subtype of tubo-ovarian carcinoma, consisting of 5% of cases. LGSC develops from serous borderline tumours (SBTs), whereas HGSC most often originates in serous tubal intraepithelial carcinoma at the fimbrial region of the fallopian tube.2 LGSC is characterised by less aggressive biological behaviour and lower sensitivity to chemotherapy compared to HGSC.3-5
In several studies, molecular analysis has shown that the mitogen-activated protein kinase (MAPK) pathway is frequently altered in LGSC, with KRAS, NRAS and BRAF mutations most often detected, while TP53 mutations, a hallmark of HGSC, are rare or absent.6-18 MAPKs are signalling proteins consisting of the extracellular signal-regulated kinases 1/2 (ERK1/2), c-Jun amino-terminal kinases 1–3 (JNK1–3), p38 (α, β, γ, and δ), and ERK5 families. They regulate critical cellular processes, including cellular development, differentiation, proliferation, and apoptosis. Ras and Raf are associated with ERK signalling and primarily mediate proliferation.19, 20
Alterations in the PI3K/AKT/mTOR signalling pathway have been reported but are relatively rare.9, 13 Mutations in other genes, as well as more extensive genomic changes, the latter including losses of chromosome domains 1p36 and 9p, have been reported.10, 13, 21 Mutations affecting the MAPK pathway have been reported to be associated with early-stage disease and/or better prognosis in some studies11, 16, 17 but not in others.12, 13
Dissemination of tubo-ovarian carcinoma within the peritoneal cavity, and less frequently the pleural cavity, in the form of malignant effusion commonly occurs, particularly with advanced-stage serous carcinoma.22 Thus, despite the fact that LGSC is an uncommon tumour, its relative incidence in serous effusions is somewhat higher. In a recent series by one of the authors of the present study (BD), LGSC was found to constitute 48/700 (6.8%) effusions.23
Previous molecular studies were performed on solid lesions, and to the best of our knowledge, there are currently no data regarding the molecular profile of LGSC in serous effusions. The objective of the present study was to investigate this issue.
2 MATERIALS AND METHODS
2.1 Patients and specimens
Effusion specimens (n = 17; 15 peritoneal, 2 pleural) were submitted to the Department of Pathology at the Norwegian Radium Hospital for routine diagnostic purposes during the period 2000–2021. Specimens were from 16 patients (1 patient with 2 peritoneal effusions), of whom 14 were diagnosed with LGSC and 2 with SBT that progressed to LGSC.
Nine surgical specimens from six patients with LGSC were studied for comparative purposes. These included three patient-matched pairs of biopsies from different anatomical sites and three single biopsies from the remaining three patients.
Effusions were centrifuged immediately after tapping, and cell pellets were frozen at −70°C in equal amounts of RPMI 1640 medium (GIBCO-Invitrogen) containing 50% fetal calf serum (PAA Laboratories GmbH) and 20% dimethylsulfoxide (Merck KGaA). Cell blocks were prepared using the thrombin clot method. Surgical specimens were frozen at −70°C without any treatment.
Effusions and surgical specimens were diagnosed based on the 2014/2020 WHO criteria by an experienced pathologist with a sub-specialty in cytopathology and gynaecological pathology (BD). For effusions, diagnosis was based on morphology in Diff-Quik-stained and Papanicolaou-stained smears, hematoxylin and eosin sections from cellblocks and immunohistochemistry, the latter consisting of PAX8, WT1 and p53 staining. Calretinin was used as a negative marker to exclude mesothelioma or reactive mesothelial cells. The pre-operative biopsy was additionally assessed in all cases with available material, and the surgical specimen was assessed in patients who received upfront surgery. Clinicopathological data are presented in Table 1.
| Patient # | Specimen type | Anatomical site | Previous chemotherapy | Age | FIGO stage | CA 125 | OS | Status |
|---|---|---|---|---|---|---|---|---|
| 1 | Effusion | Pleura | Yes | 51 | III | NA | 80 | DOD |
| 2 | Effusion | Peritoneum | Yes | 64 | IV | 2050 | 35 | DOD |
| 3 | Effusion | Peritoneum | No | 43 | III | 332 | 117 | DOD |
| 4 | Effusion | Peritoneum | No | 54 | IV | 2393 | 1 | DOD |
| 5a | Effusion | Peritoneum | No | 39 | I | NA | 120 | DOD |
| 6 | Effusion | Peritoneum | No | 46 | I | NA | 211 | NED |
| 7 | Effusion | Peritoneum | No | 60 | IV | 89 | 27 | DOD |
| Effusion | Peritoneum | Yes | ||||||
| 8 | Effusion | Peritoneum | No | 63 | III | 82 | 23 | DOD |
| 9 | Effusion | Peritoneum | No | 68 | III | 1397 | 31 | DOD |
| 10 | Effusion | Peritoneum | No | 38 | III | 1543 | 23 | DOD |
| 11 | Effusion | Pleura | Yes | 78 | IV | 367 | 41 | DOD |
| 12 | Effusion | Peritoneum | No | 60 | IV | 469 | 20 | DOD |
| 13 | Effusion | Peritoneum | No | 79 | III | 450 | 10 | DOD |
| 14 | Effusion | Peritoneum | No | 49 | III | 475 | 81 | AWD |
| 15 | Effusion | Peritoneum | No | 61 | NA | NA | 44 | AWD |
| 16a | Effusion | Peritoneum | No | 64 | NA | NA | NA | NA |
| 17 | Biopsy | Ovary | No | 52 | III | NA | 78 | DOD |
| Biopsy | Peritoneumb | No | ||||||
| 18 | Biopsy | Ovary | No | 49 | III | 478 | 133 | DOD |
| Biopsy | Peritoneumc | No | ||||||
| 19 | Biopsy | Lymph node | No | 44 | III | 102 | 218 | DOD |
| Biopsy | Peritoneum | No | ||||||
| 20 | Biopsy | Ovary | No | 31 | III | 1212 | 219 | AWD |
| 21 | Biopsy | Omentum | No | 59 | III | 1262 | 48 | DOD |
| 22 | Biopsy | Ovary | No | 82 | NA | 8 | 114 | DOC |
- Abbreviations: AWD, alive with disease; DOC, dead of other cause; DOD, dead of disease; FIGO, Federation of Gynaecology and Obstetrics (Fédération Internationale de Gynécologie et d'Obstétrique); NA, not available; NED, no evidence of disease.
- a Ovarian tumour diagnosed as serous borderline tumour.
- b Serosal surface of the colon.
- c Serosal surface of the uterus.
Informed consent was obtained according to national and institutional guidelines. Study approval was given by the Regional Committee for Medical Research Ethics in Norway (REK # S-04300).
2.2 Molecular analysis
Effusions were thawed by diluting the cells 1:10 in phosphate-buffered saline (PBS). The cell pellet was spun down at 1000 rpm for 10 minutes and the supernatant was discarded. The cell pellet was then re-suspended in 200 μl of PBS. DNA was extracted from effusions and solid tumours using the NucleoSpin Tissue, Mini kit for DNA from cells and tissue (Machery-Nagel).
DNA purity was measured using NanoDrop (Thermo Fisher Scientific). The median absorbance 260/280 ratio was 1.9 (1.8-2.1) and concentrations were determined using a Qubit fluorometer (Thermo Fisher Scientific). Targeted next-generation sequencing was performed with the Ion GeneStudio S5 system and analysed using the Ion AmpliSeq™ Cancer Hotspot Panel v2, covering 50 unique genes. The median coverage of called variants was 1991, enabling the detection of variants down to 1% allele frequency. Variants were called, annotated, and filtered with Ion Reporter Software V.5.10 (Thermo Fisher Scientific), and manually reassessed using Integrative Genomics Viewer.
2.3 Statistical analysis
Statistical analysis was performed using the SPSS-PC software package (version 28). Results were considered statistically significant for p-values < 0.05. Survival data were available for 21 of 22 patients. Overall survival (OS) was calculated from diagnosis to the date of death or last follow-up. Analysis of progression-free survival was not performed due to missing data for several patients. Univariate survival analysis was performed using the Kaplan–Meier method.
3 RESULTS
The molecular data are summarised in Table 2. Mutations were found in tumours from 14 of the 22 patients included in this study. Full concordance in the molecular profile was observed between patient-matched specimens, including both a pre- and post-chemotherapy effusion from patient #7 and the matched surgical specimens of patients #17, #18 and #19. Overall, the most common mutations were in KRAS (three patients) and BRAF (three patients), followed by NRAS (two patients), CDK2NA (two patients), TP53 (two patients) and ATM (two patients). The following genes were each mutated in only one patient: MET, STK11, ERBB2 and FLT3.
| Patient # | Mutation | Allele fraction | Type | Exon | Coding |
|---|---|---|---|---|---|
| 1 | KRAS | 77% | Missense | 3 | c.183A>C |
| 2 | No somatic variants | ||||
| 3 | No somatic variants | ||||
| 4 | No somatic variants | ||||
| 5a | MET | 35% | Missense | 14 | c.2962C>T |
| BRAF | 57% | Missense | 15 | c.1799T>A | |
| 6 | BRAF | 30% | Missense | 15 | c.1799T>A |
| 7—Specimen #1 | CDKN2A | 18% | Frameshift | 2 | c.243_244insCGCCACTCTCACCCGACCC |
| 7—Specimen #2 | CDKN2A | 29% | Frameshift | 2 | c.243_244insCGCCACTCTCACCCGACCC |
| 8 | No somatic variants | ||||
| 9 | ATM | 33% | Missense | 34 | c.5071A>C |
| TP53 | 41% | Nonsense | 7 | c.714T>A | |
| 10 | STK11 | 55% | Missense | 8 | c.1062C>G |
| 11 | No somatic variants | ||||
| 12 | No somatic variants | ||||
| 13 | ATM | 50% | Missense | 17 | c.2572T>C |
| TP53 | 11% | Missense | 6 | c.587G>C | |
| 14 | No somatic variants | ||||
| 15 | CDKN2A | 84% | Frameshift | 2 | c.211_215delAACTG |
| 16a | KRAS | 14% | Missense | c.35G>T | |
| 17—Specimen #1 | No somatic variants | ||||
| 17—Specimen #2 | No somatic variants | ||||
| 18—Specimen #1 | NRAS | 39% | Missense | 3 | c.182A>G |
| 18—Specimen #2 | NRAS | 29% | Missense | 3 | c.182A>G |
| 19—Specimen #1 | BRAF | 45% | Missense | 15 | c.1799T>A |
| 19—Specimen #2 | BRAF | 65% | Missense | 15 | c.1799T>A |
| 20 | ERBB2 | 43% | Non-frameshift insertion | 20 | c.2324_2325insATACGTGATGGC |
| 21 | KRAS | 57% | Missense | 2 | c.35G>A |
| FLT3 | 83% | Missense | 11 | c.1349C>T | |
| 22 | NRAS | 43% | Missense | 3 | c.182A>G |
- Abbreviations: NA, not available; RD, residual disease volume.
- a Ovarian tumour diagnosed as serous borderline tumour.
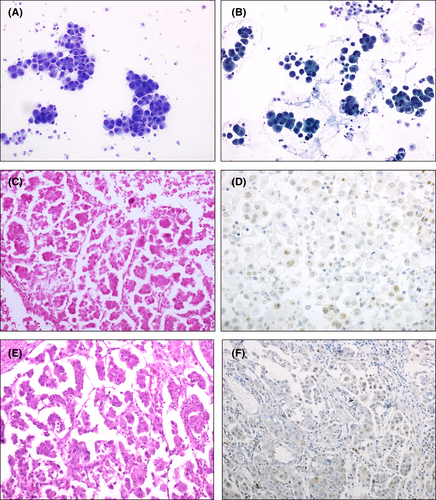
The two effusions in which LGSC cells harboured a TP53 mutation were both characterised by morphology suggestive of LGSC and stained for p53 with a wild-type pattern (Figure 1). It should be noted that ATM mutations were also found in both cases.
The number of patients with survival data in this series (n = 21) was deemed insufficient for robust analysis. However, given the reported association between mutations in genes related to the MAPK pathway and better outcomes, an attempt was nevertheless made to see whether this might be true for the current cohort. Despite the limited number of cases, patients with tumours harbouring no mutations had a considerably shorter OS than those with tumours having any mutation, though this finding was only a trend in univariate analysis (p = 0.072; Figure 2A). Dividing the tumours into three categories—those with MAPK pathway mutations, those with other mutations and those with no mutations—still generated a difference, though a statistically weaker one (p = 0.135; Figure 2B).
4 DISCUSSION
The majority of LGSC patients are diagnosed with advanced-stage disease, and despite a short-term survival advantage compared to HGSC, survival at 10 years is comparable.24 This owes much to the fact that whereas complete surgical resection results in longer survival, chemotherapy has limited effect in this disease. Given the fact that LGSC is often diagnosed in young women, considerable efforts have been made in recent years to identify molecular targets amenable to therapy, with the hope of extending survival and improving outcomes (reviewed in [3–5]). Cancer cells in serous effusions are molecularly different from their counterparts in solid lesions,22 and there is, therefore, an obvious need to characterise the genomic profile of LGSC at this anatomical site. A small series of surgical specimens was additionally studied, with an emphasis on metastatic lesions.
Analysing a case series comprising predominantly effusions, we found mutations involving the MAPK pathway in tumours from 8 of 22 patients (36%). Previous studies in which mutations in KRAS, BRAF and NRAS were all studied found a mutation in one of these genes in 37%-63% of analysed tumours,10, 12-14, 16-18—our findings are consequently at the lower end of this range. This may owe to the fact that this study focused on metastatic disease rather than on the primary tumours, thereby possibly analysing more aggressive tumour clones, having survived at extra-ovarian sites. Similar BRAF and NRAS mutations were found in patient-matched specimens in one case each.
Mutation in CDK2NA, the gene encoding p16, was found in two cases, of which one was represented by two patient-matched specimens, in agreement with the findings of a previous study by Hunter et al.,10 in which homozygous deletions of the CDKN2A/2B locus provided evidence that alterations to this gene are involved in LGSC. ERBB2 mutation, found in one case, was not found in two previous series10, 13 but was recently found in two cases of LGSC in a series of clinically aggressive SBT and LGSC cases.18 Mutations in the tyrosine kinase receptors MET and FLT3 and the kinase STK11 have not, to the best of our knowledge, been described in LGSC and therefore represent novel findings.
An unexpected finding in the present study was the presence of two tumours in which both TP53 and ATM mutations were found. TP53 mutation is a hallmark of HGSC, whereas mutations in the serine/threonine kinase ATM, a DNA damage checkpoint regulator, though less frequent, have been described in HGSC but not LGSC.25, 26 Both tumours had morphologically low-grade atypia and displayed a wild-type p53 pattern by immunohistochemistry. However, OS of 10 and 31 months from diagnosis to death-of-disease suggest that these two tumours behaved clinically like HGSC rather than LGSC.
The finding of TP53 mutations in approximately 10% of cases with an initial diagnosis of LGSC is indeed more than one would intuitively expect. However, we do not routinely perform this analysis in everyday practice, and the incidence of lack of concordance between TP53 mutation and p53 immunostaining is therefore difficult to assess. One may consider performing this analysis in cases with disseminated metastases within or outside the abdominal cavity at diagnosis. Offering PARP inhibitors to patients with tumours carrying ATM mutations seems like a reasonable treatment approach.
Assessment of the prognostic role of the detected mutations in our series is limited by its size, and larger studies which have been published in recent years are better suited to addressing this issue. We nevertheless observed a trend for better survival for patients whose tumours carried KRAS, BRAF or NRAS mutations, suggesting that observations made in some previous studies of primary LGSC11, 16, 17 may be true also for our cohort, which focused on metastatic disease. The association of a pathway mediating proliferation with longer survival may seem counter-intuitive, but this may well be related to the fact that proliferating cells are more susceptible to chemotherapy. This is in agreement with the findings of our previous study of MAPK proteins in serous carcinoma effusions, in which high expression of these molecules was associated with better outcome.27 Other mutations do not appear to be associated with the improved prognosis seen for MAPK pathway mutations based on our small series; this result may well have been affected by the two tumours with TP53 mutations.
In conclusion, the first analysis of mutation profiles in LGSC effusion shows results that are in agreement with previous studies of surgical specimens with respect to MAPK pathway mutations and their clinical relevance. We additionally report novel mutations in MET, FLT3 and STK11 in this cancer, which may provide the basis for further research and/or targeted therapy for a subset of patients.
AUTHOR CONTRIBUTIONS
Delfim Doutel: performed the laboratory work and wrote the manuscript; Ben Davidson: diagnosed the tumours, performed the statistical analysis and supervised the writing of the manuscript; Ina Kathrine Nitschke Pettersen and Annette Torgunrud: supervised the molecular analysis, interpreted the results, and critically read the manuscript.
ACKNOWLEDGEMENTS
None.
CONFLICT OF INTEREST
None declared.
Open Research
DATA AVAILABILITY STATEMENT
Research data are not shared.