Clinical and genetic spectrum in 33 Egyptian families with suspected primary ciliary dyskinesia
Funding information: Action Medical Research, Grant/Award Number: GN2101; British Council Newton-Mosharafa Fund; COST Action BEATPCD: Better Evidence to Advance Therapeutic options for PCD network (BM1407); Daniel Turnberg Travel Fellowship; Great Ormond Street Children's Charity; Ministry of Higher Education in Egypt; NIHR Biomedical Research Centre at Great Ormond Street Hospital for Children NHS Foundation Trust and University College London
Abstract
Primary ciliary dyskinesia (PCD) is a rare genetic disorder of motile cilia dysfunction generally inherited as an autosomal recessive disease. Genetic testing is increasingly considered an early step in the PCD diagnostic workflow. We used targeted panel next-generation sequencing (NGS) for genetic screening of 33 Egyptian families with clinically highly suspected PCD. All variants prioritized were Sanger confirmed in the affected individuals and correctly segregated within the family. Targeted NGS yielded a high diagnostic output (70%) with biallelic mutations identified in known PCD genes. Mutations were identified in 13 genes overall, with CCDC40 and CCDC39 the most frequently mutated genes among Egyptian patients. Most identified mutations were predicted null effect variants (79%) and not reported before (85%). This study reveals that the genetic landscape of PCD among Egyptians is highly heterogeneous, indicating that a targeted NGS approach covering multiple genes will provide a superior diagnostic yield compared to Sanger sequencing for genetic diagnosis. The high diagnostic output achieved here highlights the potential of placing genetic testing early within the diagnostic workflow for PCD, in particular in developing countries where other diagnostic tests can be less available.
1 INTRODUCTION
Primary ciliary dyskinesia (PCD, MIM#244400) is a rare inherited disorder caused by abnormal motility of cilia that is usually associated with ciliary ultrastructural defects. PCD most often manifests early in life with neonatal respiratory distress, then later with chronic respiratory disease leading to defective lung function and bronchiectasis. Affected individuals present with a range of nonspecific manifestations including chronic wet cough, rhinosinusitis, otitis media and hearing impairment. About 50% of patients have laterality problems.1 Infertility is frequently present in affected adult males.2
PCD is primarily an autosomal and X-linked recessive disease. It has high allelic and locus heterogeneity with mutations in over 40 genes known, so far, to lead to PCD (Table S1).3-5 These genes encode proteins that are either essential for the multiciliogenesis pathway or are structural and assembly proteins (cytoplasmic dynein assembly factors) of the motor machinery of the ciliary axoneme.6
In the current study, we have used targeted NGS for the genetic investigation of PCD in a cohort of Egyptian families where PCD is highly clinically suspected.
2 MATERIALS AND METHODS
2.1 Patients
The study was ethically approved by the ethics committees at the Faculty of Medicine, Alexandria, and Ain Shams Universities and the London Bloomsbury Research Ethics Committee (08/H0713/82). Forty-four patients from 33 unrelated Egyptian families were recruited based on a clinical suspicion of PCD. Self-reported consanguinity data were collected at the time of recruitment. Informed consent was obtained from all participants or their guardians.
2.2 Targeted NGS and data analysis
Targeted NGS panel of 321 genes, including all the known PCD genes and other candidate cilia motility genes, was used to screen a proband from each family (Table S1). Sequencing data were processed using an in-house bioinformatics pipeline as previously described.3, 7 A search for large insertion/deletion mutations and copy number variants was separately performed, using ExomeDepth software.8
Confirmation of the prioritized variants in the affected individuals and segregation within the available family members was performed using standard Sanger sequencing.
3 RESULTS
3.1 Clinical characteristics of the affected individuals
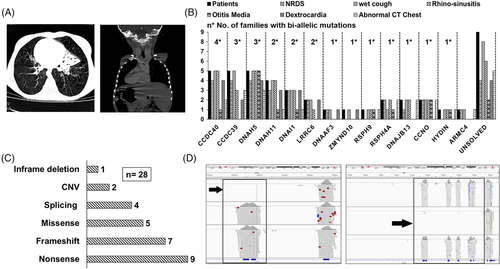
Forty-three participants were below 18 years old (1 month-18 years) at the time of recruitment, only one affected individual was 33 years old. Parental consanguinity was reported in 73% of the families. The majority of the participants showed typical PCD clinical symptoms, including a history of neonatal respiratory distress (70%), chronic wet cough (95%) and rhinosinusitis (80%). Chest CT data revealed bronchiectasis and alveolar consolidations in 76% of the examined 33 individuals. Dextrocardia was documented in more than half of the patients (55%) (Table 1). The distribution of cardiac situs according to the mutated PCD genes showed that patients with mutations in PCD genes can have either normal or abnormal cardiac situs. As expected,1 only normal cardiac situs (levocardia) was reported in patients with mutations in a specific subset of PCD genes including genes encoding central complex components (HYDIN), radial spokes components (RSPH9, RSPH4A, DNAJB13) and genes essential for multiciliogenesis (CCNO) (Figure 1A,B).
| Pa ID | Gender | Consang | NRDS | Wet cough | Rhino sinusitis | Otitis media | Cardiac situs | Chest CT | Gene | Governorate |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Male | Yes | Yes | Yes | Yes | No | Dextrocardia | Normal | CCDC40 | Alexandria |
| 2 | Male | Yes | Yes | Yes | Yes | No | Dextrocardia | Normal | - | Alexandria |
| 3 | Female | Yes | Yes | Yes | Yes | Yes | Dextrocardia | Normal | DNAH5 | Alexandria |
| 4 | Female | Yes | Yes | Yes | Yes | No | Dextrocardia | Normal | LRRC6 | Alexandria |
| 5 | Male | Yes | No | Yes | Yes | No | Dextrocardia | Not done | CCDC40 | Beheira |
| 6 | Female | Yes | No | Yes | Yes | Yes | Dextrocardia | Bronchiectasis | DNAH5 | Beheira |
| 6-a | Male | Yes | Yes | Yes | Yes | Yes | Dextrocardia | Normal | ||
| 7 | Male | No | Yes | Yes | Yes | Yes | Dextrocardia | Not done | - | Kafr Elsheikh |
| 7-a | Female | No | No | Yes | Yes | No | Dextrocardia | Not done | Kafr Elsheikh | |
| 8 | Male | Yes | Yes | Yes | No | No | Levocardia | Atelectasis | CCDC39 | Beheira |
| 8-a | Male | Yes | Yes | Yes | Yes | No | Dextrocardia | Normal | ||
| 9 | Male | No | Yes | Yes | Yes | Yes | Dextrocardia | Normal | CCDC40 | Beheira |
| 10 | Female | Yes | Yes | Yes | Yes | No | Dextrocardia | Not done | DNAH11 | Kafr Elsheikh |
| 10-a | Female | Yes | No | Yes | Yes | No | Levocardia | Bronchiectasis | Kafr Elsheikh | |
| 10-b | Male | Yes | Yes | Yes | Yes | Yes | Dextrocardia | Normal | Kafr Elsheikh | |
| 11 | Female | Yes | Yes | Yes | Yes | No | Dextrocardia | Bronchiectasis | CCDC39 | Alexandria |
| 12 | Female | Yes | No | Yes | No | No | Dextrocardia | Consolidations | ZMYND10 | Alexandria |
| 13 | Male | Yes | Yes | Yes | No | No | Dextrocardia | Not done | DNAAF3 | Cairo |
| 14 | Male | No | Yes | Yes | No | No | Levocardia | Collapse | ARMC4 | Dammietta |
| 15 | Male | Yes | Yes | Yes | Yes | Yes | Levocardia | Gas trapping | RSPH4A | Gharbia |
| 16 | Male | No | No | Yes | Yes | Yes | Levocardia | Bronchiectasis | HYDIN | Sharqia |
| 17 | Female | No | No | Yes | Yes | Yes | Levocardia | Bronchiectasis | RSPH9 | Sharqia |
| 18 | Male | Yes | Yes | Yes | No | No | Dextrocardia | Not done | LRRC6 | Gharbia |
| 19 | Female | Yes | No | Yes | No | No | Levocardia | Not done | CCDC39 | Beheira |
| 19-a | Female | Yes | Yes | Yes | Yes | No | Levocardia | Bronchiectasis | ||
| 20 | Female | Yes | No | Yes | Yes | Yes | Levocardia | Bronchiectasis | - | Alexandria |
| 21 | Female | No | Yes | Yes | Yes | No | Dextrocardia | Not done | DNAH11 | Alexandria |
| 22 | Female | Yes | Yes | Yes | Yes | No | Levocardia | Bronchiectasis | DNAJB13 | Beheira |
| 22-a | Female | Yes | No | Yes | Yes | No | Levocardia | Bronchiectasis | ||
| 23 | Female | No | No | Yes | No | No | Levocardia | Bronchiectasis | - | Alexandria |
| 24 | Male | Yes | Yes | Yes | Yes | No | Levocardia | Bronchiectasis | CCDC40 | Alexandria |
| 24-a | Male | Yes | Yes | Yes | Yes | No | Dextrocardia | Bronchiectasis | ||
| 25 | Male | Yes | Yes | Yes | Yes | No | Dextrocardia | Not done | DNAI1 | Kafr Elsheikh |
| 25-a | Female | Yes | Yes | Yes | Yes | No | Dextrocardia | Consolidations | ||
| 26 | Male | Yes | Yes | Yes | Yes | Yes | Levocardia | Bronchiectasis | CCNO | Kena |
| 26-a | Male | Yes | Yes | Yes | Yes | Yes | Levocardia | Bronchiectasis | ||
| 27 | Female | Yes | Yes | Yes | Yes | Yes | Dextrocardia | Bronchiectasis | DNAH5 | Kafr Elsheikh |
| 27-a | Male | Yes | Yes | Yes | Yes | Yes | Levocardia | Bronchiectasis | ||
| 28 | Female | Yes | No | No | No | No | Levocardia | Atelectasis | - | Alexandria |
| 29 | Male | No | Yes | Yes | Yes | Yes | Dextrocardia | Not done | DNAI1 | Alexandria |
| 30 | Male | Yes | No | Yes | No | No | Dextrocardia | Bronchiectasis | - | Beheira |
| 31 | Female | Yes | Yes | No | Yes | No | Levocardia | Bronchiectasis | RSPH4A | Beheira |
| 32 | Male | Yes | Yes | Yes | Yes | No | Levocardia | Not done | - | Alexandria |
| 33 | Male | No | Yes | Yes | Yes | No | Levocardia | Atelectasis | - | Beheira |
- Note: a and b symbols indicate the siblings of the family's proband. PCD gene shown if mutations were identified comprising biallelic variants except in the case of IDs 14 and 15, where only single heterozygous variants were identified.
- Abbreviations: NRDS, neonatal respiratory distress syndrome; Pa, proband.
3.2 Genetic landscape of PCD among Egyptian patients
Biallelic variants in 13 autosomal recessive genes were identified, Sanger-confirmed and correctly segregated in 23 families, representing a high overall genetic diagnostic output of 70%. In two families (14 and 15), only one mutated allele (single heterozygous) was identified in the known PCD genes ARMC4 and RSPH4A (Tables 1 and 2).
| ID | Gene | Zygosity | Impact | c.DNA nomenclature | Protein nomenclature | gnomAD MAF | CADD score | Ref |
|---|---|---|---|---|---|---|---|---|
| 1 | CCDC40 | Homozygous | Stop-gain | NM_017950.3: c.387C > G | NP_060420.2: p.Tyr129* | Not present | 17.95 | |
| 3 | DNAH5 | Homozygous | Missense | NM_001369.2: c.8320 T > C | NP_001360.1: p.Trp2774Arg | Not present | 29.8 | |
| 4 | LRRC6 | Homozygous | Splice donor | NM_012472.4: c.974 + 1G > A | NP_036604.2: p.? | Not present | 25.7 | |
| 5 | CCDC40 | Homozygous | Deletion | CNV (del exons 11–12) | Not present | |||
| 6 | DNAH5 | Compound Heterozygous | Frameshift | NM_001369.2: c.11258del | NP_001360.1: p.Asn3753Thrfs*5 | Not present | 36 | |
| Compound Heterozygous | Frameshift | NM_001369.2: c.2964_2965del | NP_001360.1: p.Thr990Asnfs*2 | Not present | 27 | |||
| 8 | CCDC39 | Homozygous | Stop-gain | NM_181426.1: c.2182C > T | NP_852091.1: p.Gln728* | Not present | 40 | |
| 9 | CCDC40 | Homozygous | Frameshift | NM_017950.3: c.2824_2825insCTGT | NP_060420.2: p.Arg942Thrfs*57 | 0.00003608 | 35 | |
| 10 | DNAH11 | Homozygous | Frameshift | NM_001277115.1: c.13494_13500del | NP_001264044.1: p.Ser4498Argfs*15 | Not present | 37 | |
| 11 | CCDC39 | Homozygous | Splice donor | NM_181426.1: c.210 + 2 T > C | NP_852091.1: p.? | Not present | 25.8 | |
| 12 | ZMYND10 | Homozygous | Stop-gain | NM_015896.2: c.490C > T | NP_056980.2: p.Gln164* | 0.000008029 | 24.4 | |
| 13 | DNAAF3 | Homozygous | Stop-gain | NM_001256715.1: c.48G > A | NP_001243644.1: p.Trp16* | Not present | 42 | |
| 14 | ARMC4 | Single Heterozygous | Missense | NM_018076.2: c.1706G > A | NP_060546.2: p.Arg569Gln | 0.00003585 | 25.1 | |
| 15 | RSPH4A | Single Heterozygous | Stop-gain | NM_001010892.2: c.430C > T | NP_001010892.1: p.Gln144* | 0.00001194 | 35 | 15 |
| 16 | HYDIN | Compound Heterozygous | Missense | NM_001270974.2: c.6662G > A | NP_001257903.1: p.Arg2221Gln | Not present | 24.5 | |
| Compound Heterozygous | Splice region | NM_001270974.2: c.3785 + 8C > T | NP_001257903.1: p.? | Not present | 12.71 | |||
| 17 | RSPH9 | Homozygous | Inframe del | NM_152732.4: c.804_806del | NP_689945.2: p.Lys268del | 0.0000495 | 17.86 | 16-18 |
| 18 | LRRC6 | Homozygous | Missense | NM_012472.5: c.403G > A | NP_036604.2: p.Val135Met | 0.00005641 | 23.9 | |
| 19 | CCDC39 | Homozygous | Frameshift | NM_181426.1: c.526_527del | NP_852091.1: p.Leu176Alafs*10 | 0.00001616 | 24.7 | 11 |
| 21 | DNAH11 | Homozygous | Stop-gain | NM_001277115.1: c.5845C > T | NP_001264044.1: p.Arg1949* | 0.000007101 | 42 | |
| 22 | DNAJB13 | Homozygous | Frameshift | NM_153614.3: c.623del | NP_705842.2: p.Pro208Glnfs*8 | Not present | 35 | |
| 24 | CCDC40 | Homozygous | Deletion | CNV (Deletion of exons 11–16) | ||||
| 25 | DNAI1 | Homozygous | Frameshift | NM_012144.3: c.1644del | NP_036276.1: p.Trp548* | Not present | 35 | |
| 26 | CCNO | Homozygous | Stop-gain | NM_021147.3: c.307C > T | NP_066970.3: p.Gln103* | 0.0000763 | 41 | |
| 27 | DNAH5 | Homozygous | Stop-gain | NM_001369.2: c.5034C > A | NP_001360.1: p.Cys1678* | Not present | 37 | 19 |
| 29 | DNAI1 | Compound Heterozygous | Missense | NM_012144.3: c.1352 T > A | NP_036276.1: p.Phe451Tyr | 0.000003976 | 27.6 | |
| Compound Heterozygous | Splice acceptor | NM_012144.3: c.1719-1G > A | NP_036276.1: p.? | Not present | 33 | |||
| 31 | RSPH4A | Homozygous | Stop-gain | NM_001010892.2: c.430C > T | NP_001010892.1: p.Gln144* | 0.00001194 | 35 | 15 |
The 13 mutated genes are all members of various different functional gene groups implicated in PCD.9 This therefore reveals a high genetic heterogeneity among PCD patients in Egypt, consistent with the high recorded rate of consanguineous marriage among Egyptians10 and within this study cohort (73%). The CCDC40 gene was the most frequently mutated gene within the studied cohort of families. Together, two functionally related “ruler protein” genes, CCDC40 and CCDC39,11 were the most prevalent mutated functional group of genes. Mutations in these genes result in microtubular disorganization and loss of inner dynein arms from the cilia of affected patients.
The other more prevalent mutations were detected in two genes, DNAH5 and DNAI1, encoding outer dynein arm components and associated with outer dynein arm loss. Also in DNAH11 which also codes for an outer dynein arm protein, but mutations in this gene are notably associated with normal ciliary ultrastructure when analyzed by transmission electron microscopy (TEM), as widely reported.12-14
3.3 Egyptian PCD mutation spectrum includes novel and recurrent mutations
About 79% of the variants were predicted to be null truncating variants including stop-gain, frameshift, mutations affecting splicing and CNVs. Most of the prioritized variants had a CADD score of more than 20 (Table 2 and Figure 1C). Using ExomeDepth software to search for CNVs, we identified two different homozygous deletions in CCDC40 in two unrelated families, 5 and 24 (Table 2 and Figure 1D).
Apart from a p.Gln144* stop-gain variant in RSPH4A, which was found in two unrelated families (one in a homozygous state and the other as a single heterozygous variant), all the other 26 variants were detected once per family. Interestingly, the p.Gln144* variant was reported before as a single heterozygous variant in a patient with a central microtubular complex defect.15 Three out of the remaining 26 variants were also reported before (Table 2). The inframe deletion p.Lys268del in RSPH9 has been reported several times before in Bedouin families, arising from one of the most common founder mutations in the Arab peninsula.16-19 The frameshift mutation p.Leu176Alafs*10 in CCDC39 was reported twice before in two unrelated families from Zimbabwe and UK, indicating that this is a more universal allele reported in various ethnicities.11 The stop-gain mutation p.Cys1678* in DNAH5 was also reported before, in an Italian PCD patient,20 again indicating sharing of alleles with European descent PCD patients. All the other variants identified in the current study have not been previously reported (Table 2).
4 DISCUSSION
In the current study, we have used the targeted NGS approach for genetic screening of a cohort of Egyptian patients in order to investigate the genetic landscape of PCD among Egyptians and to evaluate genetic analysis as a diagnostic tool for PCD, with high potential in developing countries that may lack an established clinical service for other specialized techniques required for diagnosis, for example, TEM and high-speed video microscopy (HSVM). The benefit of investing in genetic testing extends beyond its diagnostic value, to the appropriate genetic counseling of other family members as well as the affected individuals.
To the best of our knowledge, this is the first report to investigate the genetic background of PCD in Egypt, despite the paucity of clinical reports of Egyptian patients with PCD.21 A high genetic diagnostic output was obtained (70%). This was equivalent between patients with and without situs inversus and without any prior TEM, HSVM or nitric oxide screening; however, this cohort largely displays severe respiratory symptoms suggestive for PCD. This yield is comparable to the output of other studies, where causative mutations could be identified in 70 to 80% of PCD patients.22-25 Genetic testing has therefore proved to be a highly successful single test for PCD diagnosis in Egypt.
A 70% diagnostic success also indicates that there still remain new variants and likely also new genes to be identified to cause PCD in the Egyptian population. In two families, only a single heterozygous variant was found in affected individuals, highlighting the difficulties that can be encountered when targeted sequencing is used for genetic diagnosis of rare diseases. In these cases, a second mutation may be one that is not readily detected by this sequencing approach, such as a deep intronic mutation affecting splicing that has not been included into the NGS exome sequence capture design.26
Despite the genetic heterogeneity revealed in this Egyptian cohort, mutations in genes essential for microtubular organization (CCDC40, CCDC39) were jointly more commonly mutated than DNAH5 (considered to be the overall most commonly mutated gene in PCD27) and DNAI1 (also reportedly commonly mutated28), which both encode outer dynein arm proteins. This frequency of mutations in the two “ruler protein” genes may be due to aspects of disease recognition in Egypt, as the more severe phenotypes that are associated with mutations in these genes can more highly warrant clinicians to investigate for PCD.14 In addition to the Arabic founder mutation RSPH9 p.Lys268del,16-19 previously reported CCDC39,11 DNAH520 and RSPH4A16 mutations were detected in Egyptian patients (Table 2), probably in patients of Arabic origin but additional haplotyping could be used to establish the extent of shared ancestry outside Egypt.
In summary, we have described the use of targeted multigene panel sequencing for genetic screening of PCD in Egypt. We showed a high diagnostic yield of about 70% among patients with highly suggestive PCD diagnosis. This highlights the potential of using NGS-based genetic testing for PCD diagnosis in developing countries, as an alternative to other potentially more complex investigations, for example, cilia ultrastructural analysis by TEM.
ACKNOWLEDGEMENTS
We are very grateful to the families who participated in this study. The authors participate in the COST Action BEATPCD: Better Evidence to Advance Therapeutic options for PCD network (BM1407). M.R.F. is supported by the British Council Newton-Mosharafa Fund and the Ministry of Higher Education in Egypt. H.M. is supported by a Daniel Turnberg Travel Fellowship. Other fundings for this study were provided by Action Medical Research (GN2101; H.M.M.) and Great Ormond Street Children's Charity grant (V4515; H.M.M.) and Leadership awards (V1299, V2217; H.M.M.). We acknowledge support from the NIHR Biomedical Research Centre at Great Ormond Street Hospital for Children NHS Foundation Trust and University College London. The views expressed in this publication are those of the authors and not necessarily those of the NHS, the National Institute of Health Research, or the Department of Health.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
The supporting data are available upon request.