Ubiquitination of NOTCH2 by DTX3 suppresses the proliferation and migration of human esophageal carcinoma
Abstract
The NOTCH2 gene plays a role in the development of many tumors. Deltex E3 ubiquitin ligase 3 (DTX3) was identified as a novel E3 ligase for NOTCH2 and as a potential therapeutic target for esophageal cancer. However, whether DTX3 could regulate NOTCH2 to suppress the progression of esophageal carcinoma remains unknown. In our study, NOTCH2 had higher expression in human esophageal carcinoma cell lines compared to normal human esophageal epithelial cell line, and ablation of NOTCH2 suppressed the proliferation and migration of esophageal carcinoma cells. A novel E3 ligase for NOTCH2 was identified by yeast two-hybrid (Y2H) screening, and DTX3 promoted the ubiquitination and degradation of NOTCH2. Further study showed that DTX3 overexpression suppressed the proliferation and tumorigenicity of human oesophageal carcinoma cells. The analysis of tissue samples from patients revealed that the expression of NOTCH2 was high while the expression of DTX3 was low in esophageal cancer. Furthermore, the expression of DTX3 and NOTCH2 showed a significant negative correlation in human oesophageal cancer samples. Our study suggested that the DTX3-NOTCH2 axis plays an important role in the progression of esophageal cancer, and DTX3 acts as an anti–oncogene in esophageal carcinoma, potentially offering a therapeutic target for esophageal cancer.
1 INTRODUCTION
Esophageal cancer (EC) is a highly prevalent malignancy worldwide. According to data released by the American Cancer Society in 2018 the number of estimated new cases of EC was 17 290 in the USA, accounting for 10% of digestive system tumors; the estimated number of deaths from EC was 15 850, ranking seventh among all malignancies.1, 2 According to the data released by the National Cancer Registry Center of China in 2015, the prevalence of EC was 21.17/100 000 in China in 2012, ranking fifth among all malignancies; the mortality rate was 15.58/100 000, ranking fourth among all malignancies.3 Today, although the treatment for EC is multidisciplinary, surgery remains the most effective therapeutic approach in patients without metastatic disease.4-6 There is still no good treatment method for metastatic EC.7 Targeted therapy is widely used in cancer treatment because of its minimal side effects compared to other strategies, so new targets for EC treatment are urgently needed.
The Notch signaling pathway is implicated in self-renewal of stem cells, cell-fate determination of progenitor cells and terminal differentiation of proliferating cells.8 Notch family receptors, including NOTCH1, NOTCH2, NOTCH3 and NOTCH4, are type I transmembrane proteins with extracellular EGF-like repeats.9 DLL1, DLL3, DLL4, JAG1 and JAG2 with Delta/Serrate/LAG-2 (DSL) domain are typical Notch ligands.8 Notch-ligand binding induces the cleavage of Notch receptor by metalloprotease and γ-secretase to release Notch intracellular domain (NICD), then translocates into the nucleus to associate with DSL transcription factor which mediates gene transcription.5 Notch signaling is oncogenic in a variety of human tumors, such as through chromosomal translocation of NOTCH1 in acute lymphoblastic leukemia,10 amplification and overexpression of NOTCH2 in medulloblastoma,11 and amplification and overexpression of NOTCH3 in ovarian cancer.12 The gene mutation of Notch pathways, including NOTCH2, is reported to be involved in EC.13, 14 A notably higher NOTCH2 expression level was found in esophageal squamous cell carcinoma (ESCC) tissues, and its overexpression was significantly associated with worse overall survival and progression-free survival rates in ESCC patients.15 In this study, the function of NOTCH2 in EC cells was extensively elucidated, and deltex E3 ubiquitin ligase 3 (DTX3) was identified as a novel E3 ligase for NOTCH2 and promoted its degradation, revealing a potential therapeutic target for EC.
2 MATERIALS AND METHODS
2.1 Plasmid construction
The shRNA for NOTCH2 and DTX3 were synthesized as oligos (Sangon), annealed and inserted into the pLKO.1 vector that was digested with EcoRI and AgeI, the specific sequence for shRNA (see Table S1). The shRNA2 for NOTCH2/DTX3 were designed and targeted for the 3′-UTR sequence. The plasmids of pCDH, pCDH-NOTCH2, pCDH-DTX3-Flag, pDEST32-NOTCH2 and pDEST22-DTX3 were kindly provided by Professor Ronggui Hu (Chinese Academy of Sciences, Shanghai, China). The plasmid of pCDH-DTX3-Flag (ΔRING) was generated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene).
2.2 Cell culture, transfection and treatment
The normal human esophageal epithelial cell line (HEEC) and human esophageal carcinoma cell lines KYSE150, TE-1 and Eca-109 were cultured in RPMI-1640 medium, supplemented with 10% FBS and 100 U/mL penicillin and 100 mg/mL streptomycin (all from Gibco), in a 37°C humidified atmosphere of 5% CO2. The plasmids contained NOTCH2/DTX3 shRNA or DTX3 were transiently transfected into human esophageal carcinoma cells using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions and the stably transfected cell lines were screened by puromycin (2 μg/μL, Thermofisher). For bortezomib (BTZ), bafilomycin (BAF) or cycloheximide (CHX) treatments, cells were treated with BTZ (1 μmol/L), BAF (2 μmol/L) or CHX (100 μg/mL) for 6-8 hours before harvested, then subjected to immunoblotting analysis.
2.3 Quantitative RT-PCR
Total RNA were extracted from cells using a Total RNA Kit (Tiangen). Complementary DNA (cDNA) was synthesized using ReverTra Ace qPCR RT Master Mix (Toyobo). Quantitative PCR (qPCR) assay was performed to assess the relative abundances of NOTCH2 and GAPDH mRNA using specific primers (Table S2), stained by SYBR Green (Toyobo) on the ABI 7500 Fast Real-Time PCR System (ABI). The relative abundances of NOTCH2 were normalized to that of GAPDH gene expression, using the ΔΔCt method.16 All data were obtained from three independent experiments.
2.4 Yeast two-hybrid screen
The GAL4-based yeast two-hybrid system (Y2H, Invitrogen) was used to screen for and analyze the protein-protein interaction in yeast.16 The plasmid pDEST32-NOTCH2 containing the in-frame fusion of GAL4 DNA binding domain acted as a bait. The prey vector pDEST22 containing human cDNA collections in-frame fused to the GAL4 activating domain (Invitrogen). Using the empty pDEST22 plasmid as a negative prey control, Y2H screening was performed by transforming yeast strain (Mav203 strain) containing pDEST32-NOTCH2 with the prey vectors for the human E3 cDNA expression library. Yeast transformants were first grown on the agar plate on SD-2 (deficient in Leu, Trp) for selection of yeast cells containing both bait and prey vectors, and then transferred to SD-4 (deficient in Leu, Trp, His and Ura) plates to screen for proteins that potentially interact with human NOTCH2. Colonies grown on the SD-4 plates were picked and streaked onto another SD-4 plate with X-Gal (5-bromo-4-chloro-3-indolyl-beta-d-galactopyra-noside, Sigma) added. “Positive” colonies were sequenced after amplification in Escherichia coli. Each interaction was confirmed by transforming yeast Mav203 cells with the indicated bait and prey vectors, and allowing the transformants to grow on the SD-2 or SD-4 agar plates (with or without X-Gal) for approximately 3 days at 30°C. Images of the colonies on both plates were recorded.
2.5 Cell proliferation assay
A total of 3000 cells that stably transfected with the plasmids of pLKO.1-Scramble/pLKO.1-NOTCH2-shRNA or pCDH/pCDH-NOTCH2/pCDH-DTX3 were seeded into a 96-well-plate. Six hours after cell seeding was defined as the 0-hour time point; 0, 24, 48 or 72 hours later, the cells were incubated with CCK-8 solution (C0037, Beyotime Biotechnology) for 2 hours at 37℃; then the absorbance of cells was quantified spectrophotometrically at a wavelength of 450 nm using a Microplate Reader (Bio-rad). Experiments were finished in six replicates and repeated three times.
2.6 Colony formation assay
One thousand cells that stably transfected with the plasmids of pLKO.1-Scramble/pLKO.1-NOTCH2-shRNA or pCDH/pCDH-NOTCH2/pCDH-DTX3 were seeded into 6-well-plates. Seven days later, plates were fixed with 4% paraformaldehyde (Sigma) and stained with 0.1% crystal violet (C0121, Beyotime Biotechnology); then the colony number was counted.
2.7 Wound-healing assay
Cells were plated into 6-well plates at 105 cells/well and 100-mL tips were used to scratch the monolayer of cells when they grew to 90% confluence in each well. Then, the plates were washed by PBS twice and filled with fresh medium for 12 hours. Images of cells migrating at the wound sites were captured using an inverted microscope (IX51, Olympus).
2.8 Co–immunoprecipitation, immunoprecipitation and immunoblotting
For co–immunoprecipitation (co–IP), cells were lysed in Co–IP buffer (50 mmol/L Tris–HCl, 150 mmol/L NaCl, 5 mmol/L EDTA, 1% NP-40) supplemented with protease inhibitor cocktail (1:100, Roche), then cell lysates were incubated with anti–NOTCH2 antibody (1:100, ab245324, Abcam) and protein G agarose beads (Merck Millipore) overnight at 4°C. For immunoprecipitation, cells were lysed in IP buffer (50 mmol/L Tris–HCl, 150 mmol/L NaCl, 5 mmol/L EDTA, 0.1% SDS, 1% NP-40) supplemented with protease inhibitor cocktail after BTZ treatment (6 hours), then cell lysates were incubated with anti–NOTCH2 antibody and protein G agarose beads overnight at 4°C for Co-IP. The immunoprecipitants were enriched and denatured at 100°C for 10 minutes in 2× SDS-PAGE loading buffer. The inputs, immunoprecipitants and other cell lysates were then subjected to SDS-PAGE, and transferred to PVDF membranes (Bio-Rad). The membrane was incubated with the appropriate antibodies against NOTCH2 (1:000), DTX1 (1:1000, 18350-1-AP, Proteintech), DTX2 (1:500, 18565-1-AP, Proteintech), DTX3 (1:1000, ab116084, Abcam), DTX3L (1:800, 11963-1-AP, Proteintech), DTX4 (1:100, 25222-1-AP, Proteintech), NICH (1:600, ab8925, Abcam), ubiquitin (1:500, sc-47721, Santa Cruz), Ki-67 (1:1000, ab92742, Abcam), p-AKT (1:500, ab38449, Abcam), Bad (1:800, 10435-1-AP, Proteintech), BAX (1:1500, 60267-1-Ig, Proteintech), MMP-9 (1:1000, 10375-2-AP, Proteintech), LMNB1 (1:5000, 12987-1-AP, Proteintech), β-Tubulin (1:1000, 10094-1-AP, Proteintech) and GAPDH (1:5000, 60004-1-Ig, Proteintech). Secondary antibodies were labeled with HRP and the signals were visualized using Tanon 5200 Imaging System (Tanon).
2.9 Immunofluorescence
Cells were fixed in 4% paraformaldehyde, and incubated with anti–NOTCH2 or anti–DTX3 antibodies, followed by staining with dye-conjugated secondary antibodies and DAPI, as described previously.17 Images were taken by fluorescence microscope (BX51, Olympus) using an oil lens (100×).
2.10 Tumor xenograft assay
Five-week-old athymic female nude mice were purchased from the Shanghai Laboratory Animal Centre (SLAC). All animal studies were performed with approval from the Animal Care and Use Committee of Shanghai Changzheng Hospital, Second Military Medical University. Eca-109 cells that stably expressed. pPCDH or pCDH-DTX3-Flag were injected subcutaneously (1 × 107 cells/mouse) into the mice, with six mice each group. The volume of tumors was measured with a vernier caliper every 3 days. Mice were killed 21 days later, and the weights of tumors were measured.
2.11 Immunohistochemistry
Human EC tissue arrays were purchased from Shanghai Outdo Biotech. Immunohistochemistry for tissues from mice or patients with EC was performed as previously reported.16 Briefly, tissues were fixed in formalin, embedded in paraffin and sectioned before being mounted on slides, which were then subjected to deparaffinization and rehydration. Then, the slides were microwaved for 30 minutes in 0.01 mol/L sodium citrate buffer (PH 6.0). After antigen retrieval and pre–incubation with 10% normal goat serum, anti–DTX3 (1:50, ab116084, Abcam), anti–NOTCH2 (1:80, ab245324, Abcam), anti–Ki-67 (1:100, ab92742, Abcam) or anti–PCNA (1:100, ab29, Abcam) was used at 4℃ overnight. These slides were stained by means of the VECTSDTSIN Elite ABC Kit (Vector Laboratories) and counterstained with hematoxylin. The density of positive staining was measured using a computerized image system (Leica Microsystems Imaging Solutions). Photographs of three representative fields were captured using Leica Win Plus v3 software under high-power magnification (100×), and identical settings were used for each photograph. The density was counted using Image-Pro Plus 6.0 software (Media Cybernetics). A uniform setting for all the slides was applied for the reading of each antibody staining. The integrated optical density of all the positive staining in each photograph was measured, and its ratio to total area of each photograph was calculated as the density.18
2.12 Nuclear and cytoplasmic extract
The nuclear and cytoplasmic extracts were prepared from Eca-109 cells using a Nuclear and Cytoplasmic Protein Extraction Kit (P0027, Beyotime Biotechnology) according to the manufacturer’s instructions.
2.13 Statistical analysis
The data were analyzed by two-tailed Student’s t test or one-way ANOVA when appropriate. *P < 0.05 was considered a significant difference and **P < 0.01 was considered a very significant difference. All statistical analyses were performed using the SPSS 17.0 statistical software (SPSS) or the GraphPad Prism 5 software (GraphPad Software).
3 RESULTS
3.1 Higher expression of NOTCH2 in human esophageal carcinoma cells
To explore the expression profile of NOTCH2 in human esophageal carcinoma cells, lysates of three human esophageal carcinoma cell lines (KYSE150, TE-1 and Eca-109) and one normal HEEC were prepared. Immunoblotting analysis indicated that the protein levels of NOTCH2 were significantly higher in the three human esophageal carcinoma cell lines compared to the normal cell line, and this was consistent with the mRNA levels of NOTCH2 detected by quantitative RT-PCR (qRT-PCR) (Figure 1A). These results suggest higher NOTCH2expression in human esophageal carcinoma cells.
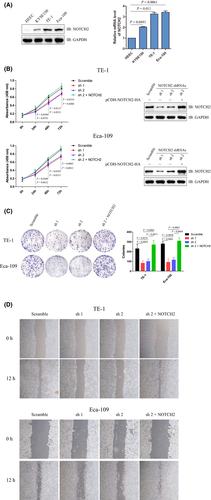
3.2 Ablation of NOTCH2 inhibits the proliferation and migration of human esophageal carcinoma cells
To explore the effect of NOTCH2 on the proliferation of esophageal carcinoma cells, shRNA for NOTCH2 were designed and tested in two esophageal carcinoma cell lines (TE-1 and Eca-109). Immunoblotting analysis indicated that NOTCH2 significantly decreased in cells stably transfected with shRNA compared to Scramble groups. Cell viability were detected by CCK-8 assay at 24, 48 and 72 hours post–culture, and obvious cell growth arrest was observed in NOTCH2 knockdown cell lines compared to control groups, while reintroduction of NOTCH2 led to a reverse of this change (Figure 1B). Next, colony formation assay was performed. Significantly decreased colony numbers were found in NOTCH2 knockdown groups compared to control groups both in TE-1 and Eca-109 cell lines, while reintroduction of NOTCH2 could reverse this phenomenon (Figure 1C). Then, the wound-healing assay indicated that NOTCH2 knockdown cells maintained longer widths 12 hours after scratching as compared to the control groups which exhibited much smaller widths, while reintroduction of NOTCH2 led to a reverse of this change, implying a poorer migration ability of cells with NOTCH2 knockdown (Figure 1D). These data suggest that ablation of NOTCH2 inhibited the proliferation and migration of human esophageal carcinoma cells.
3.3 NOTCH2 is a novel substrate for E3 ligase DTX3
The endogenous NOTCH2 protein was increased upon treatment with the proteasomal inhibitor BTZ, but not the autophagy inhibitor Bafilomycin A1 (BAF) in Eca-109 cells, suggesting that endogenous NOTCH2 protein undergoes proteasome-dependent degradation, rather than autophagy pathway (Figure 2A). Yeast two-hybrid (Y2H) screening was performed to identify the potential E3 ligase for NOTCH2. DTX3 was identified and validated in yeast cells (Figure 2B). Co-IP analysis showed that endogenous NOTCH2 could form a complex with endogenous DTX3 and DTX3L, but could not form a complex with DTX1, DTX2 and DTX4 in Eca-109 cells (Figure 2C). Immunofluorescence assay indicated that NOTCH2 and DTX3 were co–localized in the cytoplasm of human Eca-109 cells (Figure 2D). The nuclear and cytoplasmic extracts were prepared from Eca-109 cells. Immunoblotting analysis indicated that NOTCH2 was localized in the cytoplasm, while DTX3 could be detected in the nucleus and the cytoplasm, but was mainly localized in the nucleus (Figure 2E). Further immunoprecipitation analysis demonstrated that DTX3 overexpression increased the ubiquitination of NOTCH2 in Eca-109 cells (Figure 2F). Altogether, our results indicated that DTX3 interacted with and ubiquitinated NOTCH2.
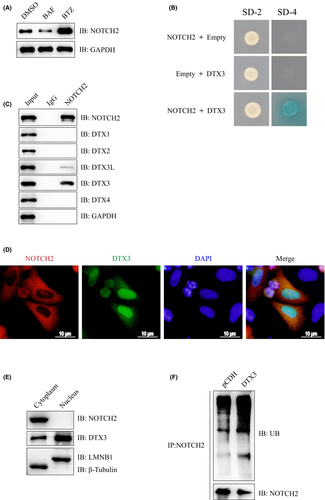
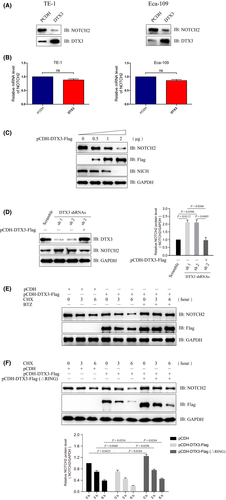
3.4 DTX3 promotes the degradation of NOTCH2
With the degradation of NOTCH2 protein in a proteasome-dependent manner (Figure 2A), we explored the effect of DTX3 on the stability of NOTCH2 protein. Immunoblotting analysis indicated that NOTCH2 significantly decreased in DTX3 overexpression groups compared to control groups both in TE-1 and Eca-109 cell lines (Figure 3A). The gene expression of NOTCH2 was detected by qRT-PCR, and we found no significant mRNA level changes in the two groups (Figure 3B). Further study indicated that DTX3 degraded the NOTCH2 protein in a dose-dependent manner (Figure 3C). The ablation of DTX3 by shRNA inhibited the degradation of NOTCH2, while reintroducion of DTX3 could rescue the protein levels of NOTCH2 (Figure 3D). Then our study found that proteasomal inhibitor BTZ could prevent the DTX3-mediated degradation of NOTCH2 protein (Figure 3E), and degradation of NOTCH2 by DTX3 depends on its E3 ligase activity (Figure 3F).
3.5 DTX3 inhibits the proliferation of human esophageal carcinoma cells
DTX3 could reduce the stability of NOTCH2 protein and its effect on the proliferation of EC cells was detected. Cell viability of TE-1 and Eca-109 was detected by CCK-8 assay, and obvious cell growth arrest was observed in DTX3 overexpression cells compared to control groups (Figure 4A). A colony formation assay was also performed, and significantly decreased colony numbers were found in DTX3 overexpression groups compared to control groups both inTE-1 and Eca-109 cell lines (Figure 4B). Then, the proliferation and apoptosis-related molecular markers were detected in Eca-109 cells that stably expressed Scramble or NOTCH2 shRNA1. Immunoblotting analysis indicated that cell proliferation-related molecules Ki-67 and p-AKT were obviously downregulated in the pCDH-DTX3 group compared to the pCDH group in Eca-109 cells that stably expressed Scramble but not NOTCH2 shRNA, while cell apoptosis-related molecules Bad and BAX were significantly upregulated in the pCDH-DTX3 group compared to the pCDH group in Eca-109 cells that stably expressed Scramble but not NOTCH2 shRNA (Figure 4C). These data suggested that DTX3 inhibits the proliferation of human esophageal carcinoma cells in a NOTCH2-dependent manner.

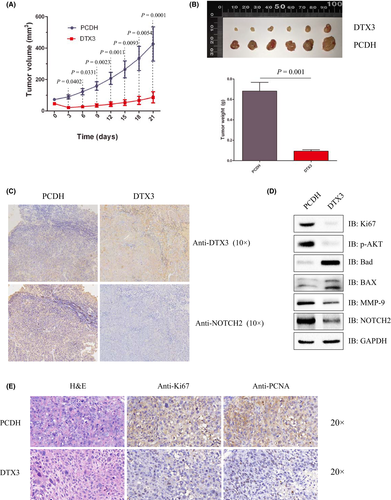
3.6 DTX3 suppresses the tumorigenicity of human esophageal carcinoma cells in nude mice
A tumorigenicity model in nude mice was used to further probe the effect of DTX3 on human esophageal carcinoma cells. Eca-109 cells that stably transfected with pCDH or pCDH-DTX3 were injected into nude mice. The tumors of the pCDH group grew quickly compared to the pCDH-DTX3 group, and the tumors in the PCDH group were significantly heavier than in the pCDH-DTX3 group (Figure 5A,B). Immunohistochemistry showed that the protein level of NOTCH2 was lower in the pCDH-DTX3 group compared to the pCDH group (Figure 5C). The tumors of nude mice were homogenized and subjected to immunoblotting analysis. The results indicated that proliferation-related molecules Ki-67 and p-AKT and migration-related molecules MMP-9 were obviously downregulated in the PCDH-DTX3 group compared to the pCDH group, while apoptosis-related molecules Bad and BAX were significantly upregulated in the pCDH-DTX3 group compared to the pCDH group (Figure 5D). Immunohistochemistry showed that proliferation-associated marker molecules Ki-67 and PCNA were lightly stained in the pCDH-DTX3 group compared to the pCDH group (Figure 5E).
3.7 The low expression of NOTCH2 while high expression of DTX3 is beneficial to patients with esophageal cancer
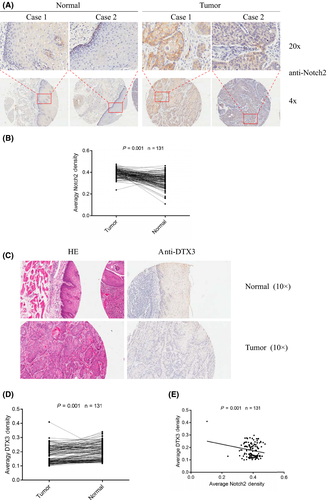
Tissues from patients with human EC were immunostained with anti–NOTCH2 or anti–DTX3 antibody, respectively. The expression of NOTCH2 was higher in cancers compared to adjacent tissues (Figure 6A,B). However, immunohistochemistry showed that the protein levels of DTX3 were lower in cancers compared to adjacent tissues (Figure 6C,D). Correlation analysis revealed that the expression of DTX3 and NOTCH2 had a significant negative correlation in human EC samples (Figure 6E).
4 DISCUSSION
The Notch signaling pathway has been extensively studied and is involved in many biology functions, such as cell proliferation, differentiation, apoptosis, adhesion, embryonic development, angiogenesis and tumor formation.19, 20 Mammalian Notch receptors include NOTCH1, NOTCH2, NOTCH3 and NOTCH4.8 NOTCH2 plays an important role in the development of various tumors, it can promote the proliferation of hepatocellular carcinoma and reduce the sensitivity of tumors to 5-fluorouracil.21 study Liu et al (2016) reported that high Notch2 mRNA expression predicted better overall survival in lung adenocarcinoma.22 However, in our study, we found that low expression of NOTCH2 is beneficial to patients with EC. Wang’s study found a notably higher NOTCH2 expression level in ESCC tissues, and its overexpression was significantly associated with worse overall survival and progression-free survival rates in ESCC patients.15 Whether NOTCH2 acts as an anti–oncogene or oncogene might depend on the cancer type. The DTX protein family has been reported to be involved in the Notch signaling pathway, and human DTX1 includes a basic N terminus, which binds the ankyrin repeats of intracellular Notch.23-25 In the fly, DTX augments Notch signals that are dependent on the downstream transcription factor Su (H).26 The ectopic DTX perturbs myogenesis, neurogenesis and lymphogenesis, and alters the transcriptional activity of E2A in mammalian systems.27-29 In our study, overexpression of DTX3 in human esophageal carcinoma cells inhibits its proliferation. The ubiquitin-proteasome pathway plays an important role in biological function.16 DTX3 contains a typical RING-type E3 ubiquitin ligase domain23 and no substrate has been reported before. In our study, DTX3 was identified as a novel E3 ligase for NOTCH2, and promoted its degradation. The protein expression of DTX3 and NOTCH2 is negatively correlated in patients with EC.
In summary, our study suggested that the DTX3-NOTCH2 axis plays an important role in the progression of EC, and DTX3 acts as an anti–oncogene in esophageal carcinoma, which might offer a potential therapeutic target for EC. An ongoing goal in our studies is to develop drugs that activate DTX3, and we believe that activating the expression of DTX3 might be a good approach for anticancer therapy.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (81402449).
DISCLOSURE
The authors declare no conflict of interest.




