Circulating exosomes contain protein biomarkers of metastatic non-small-cell lung cancer
Funding information
The National Natural Science Foundation of China (Grant/Award Numbers: ‘81773237’, ‘81672104’), the Shandong Provincial key research and development program (Grant/Award Numbers: ‘2017GSF18183’, ‘2017CXGC1207’) and the Medicine and Health Science Technology development program of Shandong province (Grant/Award Number: ‘2017WS001’).
Abstract
The present study aimed to investigate the overall changes in exosomal proteomes in metastatic and non-metastatic non-small-cell lung cancers (NSCLC) and healthy human serum samples, and evaluate the potential of serum exosomal biomarkers to predict NSCLC metastasis. Tandem mass tags combined with multidimensional liquid chromatography and mass spectrometry analysis were used for screening the proteomic profiles of serum samples. Quantitative proteome, significant pathway, and functional categories of patients with metastatic and non-metastatic NSCLC and healthy donors were investigated. In total, 552 proteins of the 628 protein groups identified were quantified. Bioinformatics analysis indicated that quantifiable proteins were mainly involved in multiple biological functions, metastasis-related pathways. Moreover, lipopolysaccharide-binding proteins (LBP) in the exosomes were found to be well distinguished between patients with metastatic and patients with non-metastatic NSCLC. Area under the curve (AUC) was 0.803 with a sensitivity of 83.1% and a specificity of 67% (P < .0001). Circulating LBP were also well distinguishable between metastatic and non-metastatic NSCLC, the AUC was 0.683 with a sensitivity of 79.5% and a specificity of 47.2% (P = .005). This novel study provided a reference proteome map for metastatic NSCLC. Patients with metastatic and non-metastatic NSCLC differed in exosome-related proteins in the serum. LBP might be promising and effective candidates of metastatic NSCLC.
Abbreviations
-
- AC
-
- adenocarcinoma
-
- EGFR
-
- epidermal growth factor receptor
-
- EV
-
- extracellular vesicle
-
- LBP
-
- lipopolysaccharide binding protein
-
- LC
-
- liquid chromatography
-
- MS
-
- mass spectrometry
-
- NSCLC
-
- non-small-cell lung cancer
-
- SCC
-
- squamous cell carcinoma
-
- TDE
-
- tumor-derived exosome
1 INTRODUCTION
Non-small cell lung cancer (NSCLC), the most common type of lung cancer that accounts for approximately 85% of all lung cancer,1 is histopathologically divided into several subtypes, including squamous cell carcinoma (SCC) and adenocarcinoma (AC).2 SCC accounts for 20%-30% of all lung cancer cases, more common in elderly men and correlated with smoking.3 AC is more frequently observed in female patients and is not correlated with smoking. Despite continuous improvements in therapeutic methods, NSCLC patients remain extremely vulnerable to relapse and mortality, and the overall 5-year survival rate still remains at 10%-15% as a result of the delay in diagnosing the disease.4 Still, diagnosing NSCLC can be challenging, and new methods to support the already used clinical tools are needed to secure a better overall outcome.5 Recently, this field has been enlivened with the exciting possibility that a newly described mode of intercellular cross-talk mediated by exosomes could have important and multifarious roles in local and distant failures, hence opening new possibilities for diagnostic, predictive approaches.6
Exosomes are secreted membrane-enclosed vesicles (extracellular vesicles) of 50-150 nm in diameter.7 They are formed during the inward budding of late endosomes, develop into intracellular multivesicular endosomes, and contain functional proteins, RNA molecules, and lipids.8-10 They are released by cells and participate in cell-to-cell communication.11 Exosome-enriched proteins include members of the tetraspanin family (CD9, CD63, and CD81), members of the endosomal sorting complexes required for transport (TSG101 and Alix), and heat-shock proteins (Hsp60, Hsp70, and Hsp90).12
Recently, TDE have been shown to promote cancer progression, allowing the distal organ site to be prepared as a premetastatic niche.13, 14 Researchers have proposed that TDE provide autocrine and paracrine signals within the tumor ecosystem to activate an epithelial-mesenchymal transition program in neoplastic epithelial cells.15-18 Evidence that exosomes may bias metastatic efficiency to different target organs derives from exosomes’ avidity for specific recipient cells.6, 19-21 Moreover, TDE are readily available in blood samples where they constitute potential biomarkers of tumor.5 TDE contain several tumor-associated proteins, such as EGFR, KRAS, and RAB-family proteins.22 Exosomal proteins may well reflect pathological processes associated with disease.23 Isolation of exosomes and their use in the clinic may substitute invasive procedures for diagnosis or the follow up of cancer patients, mainly in NSCLC, where the availability of primary tumor tissue is difficult in most patients. In addition, exosomal proteins are protected from proteinase-dependent degradation and thus can be stably detected in the circulating plasma and serum, making them ideal biomarkers for a number of clinical applications.24, 25 Thus, the present study hypothesized that exosomal proteins are potential diagnostic markers in metastatic NSCLC. To date, proteomic changes of exosomes in body fluid have been described in many malignancies.26 However, the exosome proteomics of metastatic NSCLC and specific markers from exosomes that distinguish metastatic NSCLC from non-metastatic NSCLC remain less understood.
2 MATERIALS AND METHODS
2.1 Patients and clinical samples
A total of 183 patients with NSCLC (113 males and 70 females; mean age, 58.7 years; range 34 to 88) were enrolled in the Shandong Cancer Hospital. Written informed consent was obtained from all subjects. Based on the tumor and metastasis classification system, 94 patients with non-metastatic NSCLC and 89 patients with metastatic NSCLC were analyzed. None of the patients had received any treatment. Controls were recruited from outpatients who had undergone a general medical examination. Blood samples of included patients were collected and centrifuged at 3000 g, at 4°C for 15 min. The supernatant was stored at −80°C.
2.2 Isolation of exosomal proteins from human serum
Serum samples were thawed on ice,27 and the serum was centrifuged at 10 000 g at 4°C for 30 min. Then, the samples were ultracentrifuged (Beckman Coulter Class H, R, and S preparative ultracentrifuges, Type 50.4 Ti Rotor; Beckman Coulter, Brea, CA, USA) at 100 000 g at 4°C for 120 min. Next, the exosome pellets were washed with PBS, which was filtered through a 0.22-μm pore filter, followed by a second step of ultracentrifugation at 100 000 g at 4°C for 120 min. Measurement of nanoparticle size was based on tunable resistive pulse sensing (TRPS) and conducted using the qNano (Izon Science Ltd, Christchurch, New Zealand), combining tunable nanopores with proprietary data capture and analysis software, Izon Control Suite v.3.3.2.2000 (Izon Science Ltd). The supernatant was discarded, and pelleted exosomes were resuspended in 100 μL lysis buffer (8 mol/L urea, 1% protease inhibitor, 3 μM Trichostatin A (TSA), 50 mmol/L nicotinamide (NAM), and 2 mmol/L EDTA) for proteomic analysis.
2.3 Sample preparation, MS, and interpretation of mass spectra
Protein concentration in the supernatant was determined using a 2D Quant kit (GE Healthcare, Little Chalfont, UK) according to the manufacturer's instructions. For digestion, the protein solution was reduced with 5 mmol/L dithiothreito (lDTT) (Sigma Chemical Co., St Louis, MO, USA) for 30 min at 56°C and alkylated with 11 mmol/L iodoacetamide (IAA) for 15 min at room temperature in the dark; the protein sample was then diluted for trypsin digestion. Then, the peptides were combined into 18 fractions and dried by vacuum centrifuging. The peptides were subjected to an nitrogen solubility index (NSI) source followed by tandem mass spectrometry (MS/MS) in Q Exactive (Thermo Fisher Scientific, San Jose, CA, USA) coupled online to an ultra-performance liquid chromatography (UPLC) system. The resulting MS/MS data were processed using MaxQuant with integrated Andromeda search engine (v.1.5.2.8). Gene Ontology, or GO, is a major bioinformatics initiative to unify the representation of gene and gene product attributes across all species. The GO annotation proteome was derived from the UniProt-GOA database (www. http://www.ebi.ac.uk/GOA/). The Kyoto Encyclopedia of Genes and Genomes (KEGG) connects known information on molecular interaction networks. These pathways were classified into hierarchical categories according to the KEGG website.
2.4 Immunoblotting
Lysis buffer (100 μL; 1:10 dilution: 20 mmol/L Tris [pH 7.5], 150 mmol/L NaCl, 1% Triton X-100, sodium pyrophosphate, β-glycerophosphate, EDTA, Na3VO4, leupeptin) was added to the samples, which was subsequently mixed and incubated on ice for 30 min. The denatured protein was stored in a refrigerator at −20°C. Proteins (10 μL) from EV were loaded on an 8%-15% SDS gel for electrophoresis. Primary antibodies used were rabbit polyclonal anti-HSP70 (clone W70) and rabbit polyclonal anti-LBP (both from Proteintech, Chicago, IL, USA). Peroxidase-conjugated AffiniPure goat anti-mouse (H + L) and goat anti-rabbit (H + L) (both from Jackson ImmunoResearch, West Grove, PA, USA) were used as secondary antibodies.
2.5 Enzyme-linked immunosorbent assay
Blood plasma samples of patients with metastatic NSCLC, patients with early NSCLC (stage I to IIIa), and healthy donors were analyzed for expression of LBP using a Human LBP DuoSet ELISA (R&D Systems, Minneapolis, MN, USA). For ELISA testing, all serum samples were diluted with reagent diluent (1:1000) before ELISA analysis. The samples were analyzed as recommended by the manufacturer. Protein content of exosomes was determined using a fluorescent microplate reader (Molecular Devices, Biberach an der Riß, Germany), following the manual instructions.
2.6 Statistical analysis
Data were expressed as the median with interquartile range. The data between two groups were compared using the Mann-Whitney U-test and t test. Values of P < .05 were considered to be statistically significant. All statistical analyses were carried out using SPSS 22.0 statistical software.
3 RESULTS
3.1 Isolation and identification of exosomes
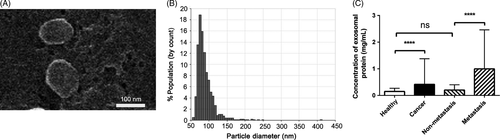
Extracellular vesicles from the serum of patients with NSCLC and healthy donors were isolated by ultracentrifugation, and were called exosomes based on the following observations. Scanning electron microscopy (SEM) showed oval-shaped extracellular vesicles of 50-150 nm diameter (Figure 1A). Moreover, the qNano system also showed that the diameter of the exosomes was 50-150 nm (Figure 1B). Quantitative results of protein concentration showed that the expression of total proteins from exosomes in patients with NSCLC was higher compared with that of healthy donors (P < .0001; Figure 1C). Median expression of proteins in patients with NSCLC was 0.416 mg/mL (range, 0.164-1.377 mg/mL) and the median expression of exosomal LBP in healthy donors was 0.234 mg/mL (range, 0.089-0.269 mg/mL). Total proteins from exosomes in patients with metastatic NSCLC were significantly higher compared with those in patients with non-metastatic NSCLC (P < .0001). Median expression of proteins was 0.996 mg/mL (range, 0.435-2.462 mg/mL) and 0.204 mg/mL (range, 0.056-0.400 mg/mL), respectively. However, no significant differences were found between healthy controls and non-metastatic NSCLC (P = .308, Figure 1C).
3.2 Profile of proteome in NSCLC cell-derived exosomes
Non-small-cell lung cancer exosomes are enriched in proteins and are implicated in the tumor microenvironment. Studies have shown that they can regulate tumor metastasis.6 In the present study, patients with distant metastatic disease (bone, brain etc., n = 5), non-metastatic NSCLC (stage I-IIIa, n = 5), and healthy donors (n = 5) were selected as a discovery cohort for proteomic analysis. In total, 628 proteins from exosomes were identified, among which 552 proteins were quantified. A quantitative ratio of more than 1.5 was considered upregulation, whereas that less than 0.667 was considered downregulation. All identified quantifiable proteins with increased (≥1.5 fold) and decreased (≤0.667) expression levels are listed in Tables S1–S4 (P < .05).
Compared with metastatic NSCLC, non-metastatic NSCLC induced 61 differentially expressed proteins: 15 upregulated proteins, such as serum amyloid A-2 protein; and 46 downregulated proteins, such as apolipoprotein M. Compared with healthy donors, NSCLC induced 43 differentially expressed proteins: 34 upregulated proteins and nine downregulated proteins.
3.3 Functional enrichment of differentially quantified proteins and clustering for protein group
Based on the differentially regulated proteins in exosomes derived from patients with NSCLC and healthy donors, the GO analysis tool was used to screen the classification of affected biological functions, including biological process, molecular function, and cellular component. The affected biological functions and the number of related proteins in patients with NSCLC and healthy donors as well as in patients with metastatic NSCLC and non-metastatic NSCLC were analyzed and compared. The top 5 affected biological functions and the amount of differentially expressed proteins are summarized in Tables 1 and 2 (Figures S1 and S2).
| Cancer vs Health | Up classification | No. proteins (%) | Down classification | No. proteins (%) |
|---|---|---|---|---|
| Biological process | Cellular process | 34 (12) | Metabolic process | 7 (16) |
| Single-organism process | 33 (11) | Biological regulation | 6 (14) | |
| Response to stimulus | 31 (11) | Single-organism process | 5 (12) | |
| Biological regulation | 30 (10) | Response to stimulus | 5 (12) | |
| Localization | 28 (10) | Cellular process | 5 (12) | |
| Molecular function | Binding | 32 (49) | Binding | 9 (53) |
| Catalytic activity | 14 (22) | Catalytic activity | 7 (41) | |
| Structural molecule activity | 6 (9) | Molecular function regulator | 1 (6) | |
| Molecular function regulator | 4 (6) | |||
| Molecular transducer activity | 4 (6) | |||
| Cellular component | Extracellular region | 33 (19) | Organelle | 9 (31) |
| Organelle | 33 (19) | Extracellular region | 7 (24) | |
| Cell | 32 (18) | Cell | 5 (17) | |
| Membrane | 22 (13) | Membrane | 3 (10) | |
| Membrane-enclosed lumen | 20 (11) | Macromolecular complex | 2 (7) |
| Metastasis vs Non-metastasis | Up classification | No. proteins (%) | Down classification | No. proteins (%) |
|---|---|---|---|---|
| Biological process | Cellular process | 14 (12) | Single-organism process | 44 (13) |
| Single-organism process | 13 (11) | Cellular process | 40 (12) | |
| Response to stimulus | 12 (10) | Biological regulation | 40 (12) | |
| Localization | 11 (10) | Localization | 33 (10) | |
| Biological regulation | 11 (10) | Response to stimulus | 32 (9) | |
| Molecular function | Binding | 13 (52) | Binding | 42 (50) |
| Catalytic activity | 5 (20) | Catalytic activity | 18 (21) | |
| Molecular function regulator | 2 (8) | Molecular function regulator | 11 (13) | |
| Chemoattractant activity | 2 (8) | Transporter activity | 6 (7) | |
| Signal transducer activity | 1 (4) | Antioxidant activity | 3 (4) | |
| Cellular component | Extracellular region | 15 (24) | Extracellular region | 43 (23) |
| Organelle | 14 (22) | Organelle | 42 (23) | |
| Cell | 11 (17) | Cell | 41 (22) | |
| Membrane | 8 (13) | Membrane-enclosed lumen | 23 (12) | |
| Membrane-enclosed lumen | 7 (11) | Membrane | 20 (11) |
The enrichment test was then carried out to identify the GO terms (molecular function, cellular component, and biological process) that were significantly enriched to indicate the nature of the differentially expressed proteins between patients with NSCLC and healthy donors. The biological process was first investigated (Figure 2A); it was found that vesicle-mediated transport [-log10 (P-value) = 3.01] was enriched with upregulated proteins. Meanwhile, the downregulated proteins related to complement activation [-log10 (P-value) = 2.39] were significantly enriched in patients with NSCLC; and on the ontology of molecular function (Figure 2B), the protein heterodimerization activity [-log10 (P-value) = 1.35] was enriched with upregulated proteins. Endopeptidase activity [-log10 (P-value) = 2.4] in downregulated proteins was enriched in patients with NSCLC. Moreover, on the ontology of cellular component (Figure 2C), the upregulated component including membrane-enclosed lumen [-log10 (P-value) = 2.43] and the downregulated proteins related to blood microparticle [-log10 (P-value) = 1.47] were significantly enriched in patients with NSCLC. The results thus indicated that biological function might be important in regulating tumorigenesis.
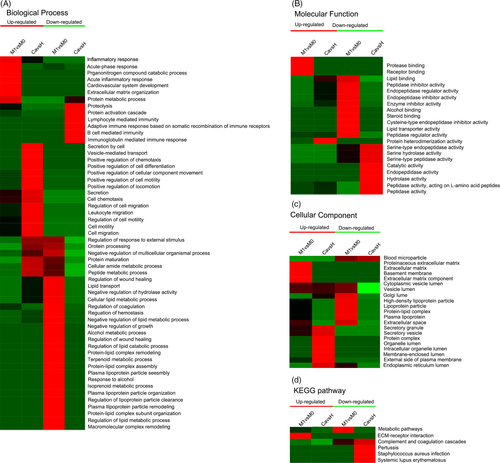
Next, enrichment tests were also carried out to identify the GO terms to indicate the nature of the differentially expressed proteins between metastatic and non-metastatic NSCLC (stage I-IIIa). On the ontology of biological process (Figure 2A), the upregulated proteins related to the acute-phase response [-log10 (P-value) = 2.7] were significantly enriched in metastatic NSCLC; meanwhile, the downregulated proteins related to protein carboxylation [-log10 (P-value) = 3.89] were enriched in biological process terms. On the ontology of molecular function (Figure 2B), protease binding [-log10 (P-value) = 2.23] was enriched with upregulated proteins and enzyme inhibitor activity [-log10 (P-value) = 2.25] was enriched with downregulated proteins. On the ontology of cellular component (Figure 2C), the upregulated component including extracellular space [-log10 (P-value) = 3.45] was significantly enriched in patients with metastatic NSCLC, and the Golgi lumen [-log10 (P-value) = 1.74] was significantly enriched with downregulated proteins. The results thus indicated that the biological function of regulating tumorigenesis and metastasis was different, and that proteome changes could distinguish between patients with metastatic and non-metastatic NSCLC. Clustering analysis based on the KEGG pathway was carried out to identify cellular pathways and the protein complex related to NSCLC (Figure 2D).
Next, the cellular pathways that related to metastatic NSCLC (Figure 3A) were identified. Results showed that the extracellular matrix-receptor interaction pathway [-log10 (P-value) = 1.81] was the dominant pathway enriched in upregulated proteins in metastatic NSCLC compared with the non-metastatic NSCLC. Meanwhile, the results showed that the metabolic pathway [-log10 (P-value) = 1.56] was the dominant pathway enriched in downregulated proteins in the same compared group.
3.4 Validation of LBP in serum exosomes could serve as a metastatic biomarker
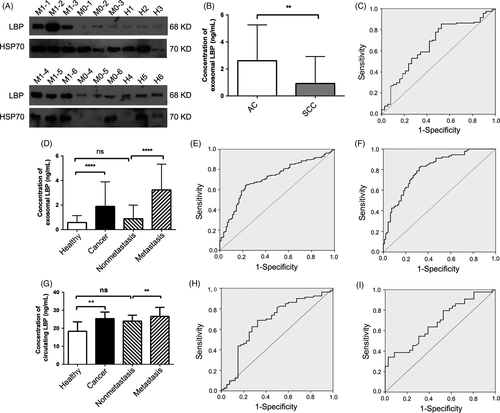
Based on the quantitative results, the list of proteins of interest was narrowed down, and functional studies were carried out for target candidates, paying more attention to the proteins with the most significant expression changes, such LBP, the fold change of which between patients with NSCLC and healthy donors was 2.574. Meanwhile, the fold change was 1.845 in metastatic NSCLC compared with the non-metastatic NSCLC.
Samples including metastatic and non-metastatic NSCLC and healthy donors were analyzed using western blotting in an independent patient cohort comprising patients with metastatic NSCLC (n = 51), patients with non-metastatic NSCLC (n = 51), and healthy donors (n = 51). Presence of exosomal marker HSP70 was confirmed by western blotting (Figure 3A). Quantitative results showed that patients with metastatic NSCLC showed high expression levels of LBP compared with patients with non-metastatic NSCLC and healthy donors. However, the non-metastatic NSCLC group showed similar exosomal LBP levels to healthy donors (Figure 3A).
Concentration of LBP was measured using ELISA in a symptomatic set with 273 subjects to determine whether LBP proteins entered the bloodstream at levels that could be measured and used as cancer biomarkers (Table3). Of the 273 individuals, 183 patients with NSCLC (89 metastatic NSCLC and 94 non-metastatic NSCLC) and 90 healthy donors were identified. Median expression level of exosomal LBP in patients with NSCLC and healthy donors was 1.895 ng/mL (range 0.588-3.887 ng/mL) and 0.577 ng/mL (range 0.143-1.138 ng/mL), respectively, with a significant difference (P < .0001; Figure 3D). Area under the curve (AUC) was 0.713 with a sensitivity of 65% and a specificity of 75.6% (Figure 3E). Based on this result, expression level of exosomal LBP from patients with metastatic and non-metastatic NSCLC was further analyzed. Analysis of the validation cohort showed that median expression level of exosomal LBP from patients with metastatic NSCLC and patients with non-metastatic NSCLC was 3.23 ng/mL (range 1.876-5.330 ng/mL) and 0.881 ng/mL (range 0.204-1.994 ng/mL), respectively. Expression level of exosomal LBP in patients with metastatic NSCLC was significantly higher than the level in those with non-metastatic NSCLC (P < .0001; Figure 3D). Receiver operating characteristic (ROC) curves indicated that exosomal LBP proteins had an AUC of 0.803 with a sensitivity of 83.1% and a specificity of 67% (Figure 3F). However, no statistically significant difference was found between patients with non-metastatic NSCLC and healthy donors (P = .09; Figure 3D).
| Characteristics | No. cases | Median with interquartile range (ng/mL) | P-value |
|---|---|---|---|
| Age (year) | |||
| 33-59 | 89 | 3.032 (0.881-5.353) | .174 |
| 60-88 | 94 | 1.947 (0.701-3.869) | |
| Gender | |||
| Male | 113 | 2.386 (0.765-5.260) | .404 |
| Female | 70 | 2.716 (0.799-3.863) | |
| Smoking status | |||
| Smoker | 102 | 2.358 (0.826-4.513) | .829 |
| Never smoked | 81 | 2.267 (0.522-5.079) | |
| Histological type | |||
| Adenocarcinoma | 117 | 2.609 (1.188-5.260) | .003 |
| Squamous cell | 38 | 0.918 (0.361-2.924) | |
| Unknown | 28 | ||
| Pathological stage | |||
| Non-metastasis | 94 | 0.881 (0.204-1.994) | <.0001 |
| Metastasis | 89 | 3.231 (1.876-5.330) | |
- LBP, lipopolysaccharide binding protein; NSCLC, non-small-cell lung cancer.
Expression level of selected exosomal LBP of patients with AC and SCC was analyzed using ELISA. Results showed that the median expression level of exosomal LBP from patients with AC and patients with SCC was 2.609 ng/mL (range 1.188-5.260 ng/mL) and 0.918 ng/mL (range 0.361-2.924 ng/mL), respectively, showing an AUC value of 0.659 with a sensitivity of 85.5% and a specificity of 47.4% in distinguishing patients with AC from patients with SCC (P = .003; Figure 3B,C). Next, serum samples of the validation cohort (40 healthy donors, 36 non-metastatic NSCLC, and 44 metastatic NSCLC) were analyzed for circulating LBP levels. Results showed that the median expression level of circulating LBP was 25.334 ng/mL (range 20.503-29.020) in patients with NSCLC and 18.340 ng/mL (range 15.643-23.569) in healthy donors, and the AUC was 0.676 with a sensitivity of 68.8% and a specificity of 67.5%. This indicated that patients with NSCLC had a high expression level of circulating LBP compared with the healthy donors (P = .002; Figure 3G,H). Moreover, high levels of circulating LBP were associated with advanced stages and presence of metastasis in metastatic NSCLC. Expression level of circulating LBP was higher in patients with metastatic NSCLC (95% confidence interval [CI], 27.964 [25.456-30.472]) compared with patients with non-metastatic NSCLC (95% CI, 22.551 [20.638-24.464]; P = .005; Figure 3G). ROC curves showed an AUC of 0.683 with a sensitivity of 79.5% and a specificity of 47.2% (Figure 3I). Moreover, the circulating LBP levels in patients with non-metastatic NSCLC were similar to those in healthy donors (P = .061; Figure 3G).
4 DISCUSSION
Exosomes carry several validated and surrogate non-invasive biomarkers with diagnostic, prognostic, and predictive values. Isolation of TDE and their use in the clinic might substitute invasive procedures for diagnosis or the follow up of NSCLC, where the availability of primary tumor tissue is difficult in most patients. So far, only a limited number of studies have examined the diagnostic potential of exosomes in relation to NSCLC. However, no studies have reported on the diagnosis of metastatic NSCLC.
At present, protein MS is highly developed and extensively used. Bioinformatics analysis further indicated that these quantifiable proteins are mainly involved in multiple biological functions and metastasis-related pathways as well as protein complexes. Furthermore, through in-depth excavation of MS data, a differentially expressed protein, LBP, was selected to determine the difference between the 3 groups. Differences in the expression of proteins might help to look for potential diagnostic and prognostic markers. Analysis of the expression of LBP in exosomes and in the circulation of patients with NSCLC showed that both could distinguish patients with NSCLC from healthy donors, but the diagnostic efficacy was not high. Further analysis showed that the level of LBP in exosomes and in the circulation was significantly different between patients with metastatic and non-metastatic NSCLC, indicating that LBP had a great potential as a biomarker for metastatic NSCLC; however, the diagnostic efficacy of circulating LBP was significantly inferior to that of exosomal LBP. Furthermore, this novel study used the proteomic technique to find a diagnostic marker for metastatic NSCLC; it provided an objective basis for the early diagnosis, early treatment, and prognosis of metastatic NSCLC, and a key point for diagnosing other cancerous solid tumors.
LBP is an acute-phase protein with a molecular weight of approximately 65 kDa, present in blood at high concentrations, and mainly produced by the liver.28-30 It has a variety of biological activities, such as anti-inflammatory effects.31 Moreover, LBP is reported to be a part of the family of lipid transfer proteins, including cholesterol ester transfer protein and phospholipid transfer protein.32 In this study, these effects were believed to be regulated by changes in lipid composition and selected phosphatidylethanolamine plasma hormones, sphingolipids, ceramides, sphingomyelin, phosphatidylserine, and glycerolipid. LBP has phospholipid transporter activity.33 No relevant study is available on the mechanism of action and changes in exosomal LBP in malignant tumors. Based on the results of the present study, exosomal LBP was speculated to be the driving factor of metastatic NSCLC. This study broadened the idea of studying the metastasis of NSCLC. Also, LBP and other differential expression proteins were likely to be important resources for finding new diagnostic biomarkers and potential targets of drugs. However, the study of proteomics of malignant tumor exoplasm is still in its infancy. According to the results of the bioinformatics analysis, the proteins that had a marked change in some specific pathways, biological functions, and developing antibodies specifically against the selected targets were then chosen to validate the selected targets biochemically. An in-depth study of exocrine proteomics will help understand the mechanism of action of human exosomes and even the whole-cell vesicle system in tumor development, leading to a better understanding of the clinical application of exosomes.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (81773237 and 81672104), the Shandong Provincial key research and development program (2017GSF18183 and 2017CXGC1207) and the Medicine and Health Science Technology development program of Shandong province (2017WS001). All authors confirm they have contributed to the intellectual content of this paper and have met the following requirement that they have significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data.
CONFLICTS OF INTEREST
Authors declare no conflicts of interest for this article.




