Downregulation of delta-aminolevulinate dehydratase is associated with poor prognosis in patients with breast cancer
Funding Information
National Natural Science Foundation of China (81372843, 81472472, 81502518); National Science and Technology Support Program (No. 2015BAI12B15), the National High Technology Research and Development Program of China (No. 2012AA021003) and Anticancer Key Technologies R & D Program of Tianjin (12ZCDZSY16200).
Abstract
Delta-aminolevulinate dehydratase (ALAD) catalyzes the second step in the biosynthesis of heme and is also an endogenous inhibitor of the 26S proteasome. The role of ALAD in breast cancer progression is still unclear. In this study, we found that the expression of ALAD was downregulated in breast cancer tissues compared with adjacent normal breast tissues. Enhanced ALAD expression was associated with a favorable outcome in patients with breast cancer. Overexpression of ALAD suppresses breast cancer cell proliferation and invasion and inhibits the epithelial–mesenchymal transition phenotype. Furthermore, we found that ALAD regulates transforming growth factor-β-mediated breast cancer progression. This finding suggests that ALAD might be a potential biomarker for breast cancer that suppresses breast cancer progression by regulating transforming growth factor-β-mediated epithelial–mesenchymal transition.
Breast cancer has been the most frequently diagnosed malignancy and it is the leading cause of cancer death in women.1 Being a heterogeneous disease, breast cancer has at least five major tumor subtypes. Different subtypes carry different risks and prognoses.2 Although we have different treatment methods for different genetic types of breast cancer, these analyses have yet to alleviate metastatic and recurrence risk.3 Up to 30% of breast cancer patients diagnosed with early stage disease suffer metastatic and recurrent process, which greatly contributes to the aggressiveness and malignant degree of the tumor.4 Thus, the molecular mechanisms of breast cancer are poorly understood and will not be discussed further here.
Epithelial–mesenchymal transition (EMT) is a complex process that leads to mammary epithelial cells (MECs) to acquire highly migratory fibroblastic-like features.5, 6 The malleability of MECs makes them dedifferentiate during EMT. In this way, transitioning MECs acquire a spindle-shaped appearance characteristic of mesenchymal cells.7 Inappropriate reactivation of EMT is associated with cell invasion and metastasis in breast cancer.8 Transforming growth factor-β (TGF-β) is a multifunctional cytokine that regulates numerous physiological processes, such as cellular differentiation, homeostasis, and EMT.9 It is a tumor suppressor that can suppress mammary tumorigenesis by preventing MEC proliferation, or by inducing MEC apoptosis.10 During mammary tumorigenesis, genetic and epigenetic events undermine the tumor suppressive functions of TGF-β, so making the cells acquire invasive and metastatic phenotypes in response to TGF-β. Recent findings have implicated TGF-β is a master regulator of EMT in normal and malignant MECs,11 and stimulate EMT with the acquisition of “stemness” in transitioning MECs,12 and with the selection and expansion of breast cancer stem cells. In brief, TGF-β has the power to induce EMT that stimulates metastatic progression in breast cancer.13
Delta-aminolevulinate dehydratase (ALAD) is encoded by the ALAD gene. It catalyzes the second step in the biosynthesis of heme and is also an endogenous inhibitor of the 26S proteasome. Recent research has suggested that ALAD and lead exposure related to cancer risk.14, 15 A number of studies indicate ALAD variants related to risk of the renal cell carcinoma,16 prostate cancer,17 and brain tumors.18 There have been no reports regarding the correlation between ALAD and breast cancer, until now. In our study, we found that downregulated ALAD is associated with a mesenchymal phenotype and poor prognosis in breast cancer that could regulate the TGF-β-mediated EMT in breast cancer cells. Although the research is not definitive about the molecular mechanisms on EMT mediating the initiation and resolution of breast cancer metastasis, our study suggested that ALAD could inhibit the mesenchymal phenotype and suppress breast cancer progression by its involvement in TGF-β-induced EMT.
Materials and Methods
Cell culture and clinical samples
MCF10A, MCF7, T47D, BT474, MDA-MB-468, MDA-MB-231 and MDA-MB-435 cell lines were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured as previously described.19 Cells were used within 6 months of resuscitation.
A total of 188 primary breast cancer specimens and 30 cases of adjacent histological normal tissues were obtained from patients who underwent breast surgery in Tianjin Medical University Cancer Institute and Hospital (Tianjin, China). All tumors were from patients with newly diagnosed breast cancer who had received no therapy before sample collection. After radical mastectomy, the primary breast cancer tissue and adjacent normal tissues were flash-frozen in liquid nitrogen and stored at −80°C. This study was approved by the Institutional Review Board of the Tianjin Medical University Cancer Institute and Hospital and written consent was obtained from all participants.
Antibodies, reagents, plasmids, and siRNAs
Antibodies against N-cadherin, E-cadherin, vimentin, ZEB1, (Santa Cruz Biotechnology, Santa Cruz, CA, USA), Snail (Abcam, Cambridge, MA, USA), and β-actin (Cell Signaling Technology, Beverly, MA, USA) were used. Recombinant human TGF-β1 was purchased from R&D Systems (Redmond, WA, USA). The ORF of the human ALAD gene was amplified by PCR in the MCF10A cell line, and the amplified fragments were subcloned into the pcDNA3.1-HA vector. The ALAD gene-specific siRNA and non-specific control siRNA were purchased from RiboBio (Guangzhou, China).
RNA extraction and RT–quantitative PCR
The total RNA of cultured cells and surgically resected fresh breast tissues were extracted using mirVana PARIS kit (Life Technologies, Gradn Island, NY, USA) according to the manufacturer's instruction. Reverse transcription was performed using a First Strand cDNA Synthesis kit (TaKaRa, Dalian, China), according to the manufacturer's instruction. The real-time quantitative PCR (qPCR) was carried out using GoTaq qPCR Master Mix (Promega, Madison, WI, USA) on a Bio-Rad iQ5 Optical System (Bio-Rad, Richmond, CA, USA). β-Actin was used as an internal control.
Western blot analysis, immunofluorescence, and immunohistochemistry
Total protein was extracted by lysing the cells with RIPA buffer and protease inhibitor. After denaturing, proteins were run in 10% SDS-PAGE gel and transferred to PVDF membranes. Membranes were blocked in 5% skim milk for 1 h at room temperature. Primary antibody was incubated overnight at 4°C. After washing in TBST, membranes were incubated with secondary antibody at room temperature for 1 h. Protein bands were visualized by the ECL system (Millipore, Bedford, MA, USA).
For immunofluorescence assay, cells were seeded in a 24-well plate. The attached cells were fixed by 4% paraformaldehyde for 30 min, and penetrated by 0.5% Triton X-100 for 15 min, then blocked by 3% BSA for 1 h. The cells were then incubated with primary antibody in 1% BSA overnight at 4°C. After washed with PBS, the cells was incubated with fluorescein isothiocyanate or phycoerythrin-conjugated secondary antibodies at room temperature for 1 h and then stained with DAPI. Finally, coverslips were observed under a fluorescence microscope.
Expression of ALAD, E-cadherin, and vimentin in breast cancer tissues was detected by immunohistochemical staining as previously described20 and were scored as negative and positive using 20% positive staining cut-off. The specimens were observed by two pathologists.
Transforming growth factor-β signaling analysis
The TGFβ signaling activity was determined by Cignal SMAD Reporter Assay Kits (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The SMAD reporter is a mixture of an inducible SMAD-responsive luciferase construct and a constitutively expressing Renilla construct. TGFβ signaling activity was detected by using a dual luciferase assay kit (Promega) according to the manufacturer's recommendations. The results were normalized against Renilla luciferase activity. All transfections were performed in triplicate.
Apoptosis assay
The cells were collected after 48 h of transfection. The cell apoptosis ratio was analyzed using an annexin V–FITC apoptosis detection kit (BD Biosciences, San Diego, CA, USA) according to the manufacturer's instructions. Apoptosis cells were analyzed by FACS.
Proliferation and invasion assays
Both MTT and plate colony formation assays were used to evaluate the ability of cell proliferation. For MTT assay, 24 h after transfection, 5 × 103 cells/well were seeded in 96-well plates. After incubation for the indicated time, the cells were incubated with 10 μL MTT (0.5 mg/mL; Sigma-Aldrich, St Louis, MO, USA) at 37°C for 4 h. Then, the medium was removed, and precipitated Formosan was dissolved in 150 μL DMSO. The absorbance at 570 nm was detected using a microplate autoreader (Bio-Rad). For the plate colony formation assay, 24 h after transfection, 500 cells/well were seeded in 6-well plates. After approximately 2 weeks, the colonies obtained were washed with PBS and fixed with 10% formalin for 15 min at room temperature, then washed with PBS and stained with hematoxylin. The colonies were counted and compared with control cells.
The invasive ability of breast cancer cells in vitro was evaluated by Matrigel-coated Transwells. Briefly, 5 × 104 cells in 500 μL serum-free medium were added to the upper chamber, and medium containing 20% FBS was added into the lower chamber. Twenty-four hours later, the migrant cells that had attached to the lower surface were fixed with 20% methanol and stained for 20 min with crystal violet. The membranes were then carved and embedded under coverslips with the cells on the top. The number of migrating cells was counted under a microscope in five predetermined fields.
Independent datasets for validation
The validation study was carried out using Gene Expression-based Outcome for Breast Cancer Online (GOBO; http://co.bmc.lu.se/gobo). This is an online tool for prognostic validation of single genes, sets of genes, or simple predictors in a pooled breast cancer dataset analyzed using Affymetrix U133A arrays (St Louis, MO, USA). Seven public datasets (accession nos. GSE11121, GSE12093, GSE2034, GSE2603, GSE5327, GSE6532, and GSE7390), which contain the DFS information, were included in the validation of prognostic value of ALAD mRNA levels.
Statistical analysis
All the experiments were carried out at least twice independently and data are presented as mean ± SEM. All statistical analyses were undertaken using spss 18.0 software for Windows (SPSS Inc., Chicago, IL, USA). For paired samples, the statistical significance of difference was calculated using Wilcoxon's signed rank with significant differences defined as at least a P-value of <0.05.
Results
ALAD is downregulated and associated with a mesenchymal phenotype in breast cancer
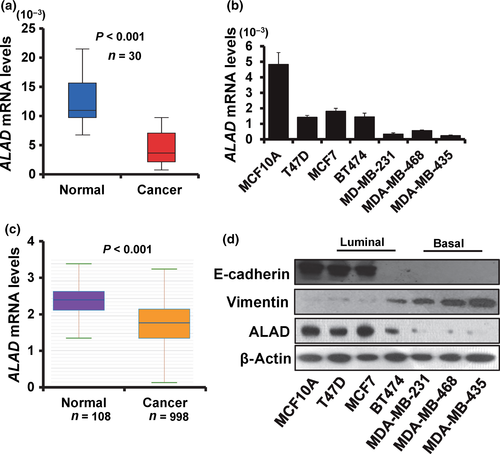
To explore the influence of ALAD on breast cancer development and progression, we first examined the expression of ALAD in 30 cases of breast cancer tissues and paired normal breast tissues by RT-qPCR. As shown in Figure 1(a), ALAD mRNA expression was downregulated in breast cancer tissues compared with adjacent normal breast tissues. To further validate the expression of ALAD in breast cancer and normal tissues, we analyzed the ALAD mRNA expression profiling dataset from the starBase version 2.0 project including 998 cases of breast cancer tissues and 108 cases of normal breast tissues.21, 22 The validation data confirmed that ALAD mRNA expression was downregulated in breast cancer tissues (Fig. 1b). We next examined the expression of ALAD in six breast cancer cell lines (T47D, MCF7, BT474, MDA-MB-231, MDA-MB-468, and MDA-MB-435) and breast epithelial cell line MCF10A by RT-qPCR and Western blot analysis. At the mRNA level, the breast epithelial cell line MCF10A showed high expression level of ALAD, whereas the basal-like breast cancer cell lines MDA-MB-231, MDA-MB-468, and MDA-MB-435 showed low expression levels of ALAD (Fig. 1c). We confirmed the expression level of ALAD at the protein level (Fig. 1d). Furthermore, the high ALAD-expressing cell lines showed high expression levels of E-cadherin and low expression levels of vimentin (Fig. 1d). These results suggested that ALAD is downregulated in breast cancer cells and low levels of ALAD expression are related to a mesenchymal phenotype.
ALAD suppresses breast cancer progression
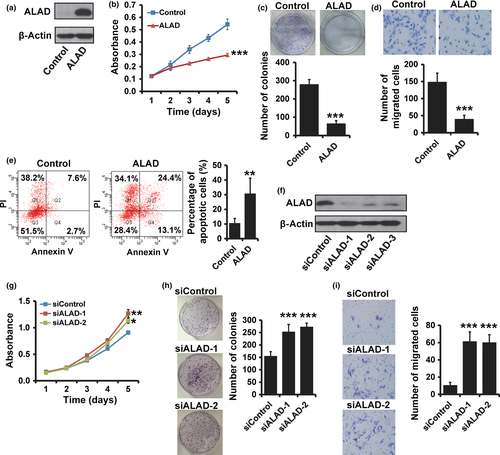
To investigate whether ALAD regulates breast cancer progression, we transiently transfected MDA-MB-231 cells with a human ALAD expression vector (Fig. 2a). We observed that cell proliferation was significantly reduced in ALAD-overexpressing MDA-MB-231 cells by MTT (Fig. 2b) and colony formation assays (Fig. 2c). Moreover, overexpression of ALAD decreased the in vitro invasion of the transfected cells (Fig. 2d). Furthermore, the number of apoptotic cells was significantly higher in ALAD-overexpressing MDA-MB-231 cells (Fig. 2e). Next, we examined the effect of the siRNA knockdown of ALAD in MCF10A cells (Fig. 2f), which normally have high ALAD expression, to determine whether depletion of ALAD affects breast cancer progression. The results from MTT (Fig. 2g) and colony formation (Fig. 2h) assays revealed that MCF10A cells transfected with ALAD siRNA showed an increase in cell proliferation compared with cells transfected with control siRNA. Invasive ability was also elevated in ALAD-depleted MCF10A cells (Fig. 2i). The same results were also observed in MDA-MB-468 and T47D cells (Fig. S1). These results suggested that ALAD suppresses breast cancer progression.
ALAD inhibits the mesenchymal phenotype in breast cancer cells
Next, we observed that overexpression of ALAD inhibited the mesenchymal spindle-like morphology in MDA-MB-231 cells (Fig. 3a). We then examined both epithelial and mesenchymal markers by RT-qPCR (Fig. 3b), Western blot analysis (Fig. 3c), and immunofluorescence (Fig. 3d). As can be seen, the ALAD-overexpressing MDA-MB-231 cells showed a significant downregulation of vimentin and N-cadherin, whereas the epithelial marker E-cadherin was dramatically upregulated. Reverse transcription–qPCR and Western blot analyses also revealed the inhibition of EMT-related factors Snail and ZEB1 in ALAD-expressing MDA-MB-231 cells (Fig. 3b,c). Thus, our study suggested that ALAD could inhibit the mesenchymal phenotype in breast cancer cells.

ALAD regulates the TGF-β-mediated EMT-like phenotype in breast cancer cells
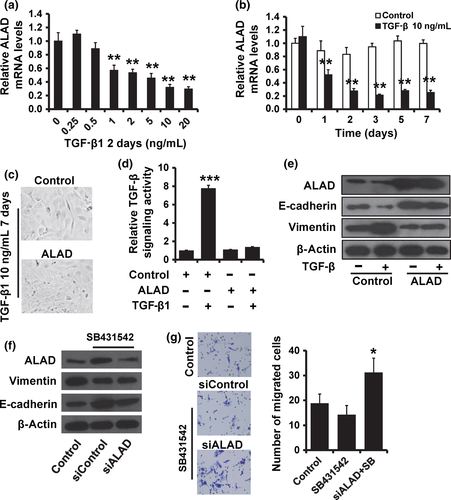
Transforming growth factor-β signaling is central to carcinogenesis and cancer progression because it regulates a variety of critical cellular processes, including EMT. Thus, we investigated the role of ALAD in TGF-β signaling. We observed that ALAD was suppressed in a dose- and time-dependent manner following the addition of TGF-β to the cell culture medium by RT-qPCR (Fig. 4a,b). To investigate whether downregulation of ALAD is required for TGF-β-induced EMT-like phenotype, we overexpressed ALAD in MCF10A cells and examined their responses to treatment with TGF-β1. MCF10A cells transfected with vector control displayed an elongated fibroblast-like morphology, whereas ALAD-transfected cells retained their cobblestone-like morphology after TGF-β1 treatment (Fig. 4c). To confirm whether TGF-β plays an important role in the oncogenic function of ALAD, we used the luciferase assay to determine the effect of ALAD on the TGF-β signaling activity. As shown in Figure 4(d), the TGF-β activity was significantly enhanced in MCF10A cells after TGF-β1 treatment. Furthermore, in MCF10A cells transfected with ALAD, the addition of TGF-β1 did not affect the TGF-β signaling (Fig. 4d). We than examined both epithelial and mesenchymal markers by Western blot analysis. Consistent with morphologic changes, E-cadherin expression was decreased, while vimentin expression was increased in MCF10A cells after TGF-β1 treatments, but neither changed expression in ALAD-expressing MCF10A cells (Fig. 4e). Our previous study indicated that TGF-β inhibitor SB431542 suppresses cancer progression in MDA-MB-231 cells.23 To further confirm the role of ALAD in TGF-β signaling, we added the TGF-β inhibitor SB431542 in ALAD-depleted MCF10A cells. We observed that the expression of E-cadherin was downregulated, whereas the expression of vimentin was upregulated, in ALAD-depleted MCF10A cells compared with that of control cells (Fig. 4f). Moreover, depletion of ALAD could partly reduce the SB431542-induced changes in MCF10A cells (Fig. 4f,g). These results suggest that ALAD is involved in TGF-β-induced EMT.
ALAD downregulation is associated with poor prognosis in patients with breast cancer
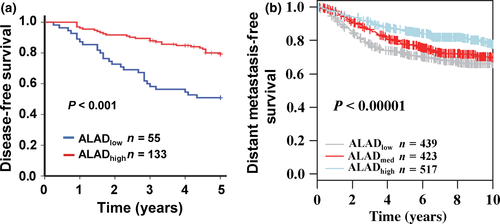
To examine whether the biological effects of ALAD on breast cancer progression were clinically relevant, we assessed its expression in 188 cases of breast cancer tissues. We then subdivided the tumors into two groups according to receiver operating characteristic curve and the optimal cut-off value was defined to classify all patients into distinguished prognosis groups (Fig. S2). The sensitivity and specificity were 0.500 and 0.707, respectively. The cut-off value 2 × 10−3 was used to group patients into low ALAD expression (ALADlow, n = 55) and high ALAD expression groups (ALADhigh, n = 133). The association between ALAD expression and clinicopathological factors are shown in Table 1. A Kaplan–Meier survival analysis revealed that patients in the ALADhigh group had a more favorable outcome than those in the ALADlow group (Fig. 5a). The result was confirmed by GOBO that patients with high ALAD expression had a favorable outcome (Fig. 5b). To confirm the in vitro finding that TGF-β-mediated mesenchymal phenotype and inhibition of ALAD are correlated in breast cancer cells, we evaluated the expression levels of ALAD, E-cadherin, and vimentin by immunochemistry in 150 breast cancer patient samples. We found that the percentage of samples with ALADneg./E-cadherinneg./vimentinpos. or ALADpos./E-cadherinpos./vimentinneg. was 50.7% (Table 2; P < 0.001).
| Clinicopathological factors | ALADhigh (n = 133) | ALADlow (n = 55) | P-value |
|---|---|---|---|
| Age, years | |||
| <55 | 84 | 40 | 0.239 |
| ≥55 | 49 | 15 | |
| Menopausal status | |||
| Pre | 61 | 33 | 0.055 |
| Post | 72 | 22 | |
| Tumor size, cm | |||
| ≤2 | 31 | 7 | 0.114 |
| >2 | 102 | 48 | |
| Clinical stage | |||
| I+II | 115 | 42 | 0.129 |
| III | 18 | 13 | |
| Histological grade | |||
| I+II | 121 | 38 | 0.001 |
| III | 12 | 17 | |
| Lymph node status | |||
| Negative | 54 | 17 | 0.249 |
| Positive | 79 | 38 | |
| ER status | |||
| Positive | 84 | 25 | 0.034 |
| Negative | 49 | 30 | |
| PR status | |||
| Positive | 68 | 25 | 0.523 |
| Negative | 65 | 30 | |
| HER-2 status | |||
| Negative | 90 | 38 | 1.000 |
| Positive | 43 | 17 | |
- ER, estrogen receptor; HER-2, human epidermal growth factor receptor-2; PR, progesterone receptor.
| Expression | ALADpos. | ALADneg. |
|---|---|---|
| E-cadherinpos. + vimentinpos. | 11 (7.3) | 15 (10.0) |
| E-cadherinneg. + vimentinneg. | 9 (6.0) | 18 (12.0) |
| E-cadherinneg. + vimentinpos. | 8 (5.3) | 55 (36.7) |
| E-cadherinpos. + vimentinneg. | 21 (14.0) | 13 (8.7) |
- Data are shown as n (%).
Discussion
Delta-aminolevulinate dehydratase is known as an important rate-limiting enzyme in the porphyrin and heme biosynthetic pathway. It has been found to play a critical role in lead metabolism and susceptibility to lead toxicity. Report of the relation between ALAD and carcinogenesis is limited. We observed the low expression of ALAD in breast cancer tissues and cell lines, both at gene level and protein level, which suggested a tumor suppressor role in breast cancer progression. To confirm that effect, human ALAD expression vectors were transfected into breast cancer cell lines, leading to the reduction of proliferation and invasion capability. In addition, the ability of proliferation and invasion was elevated in breast epithelial cell line MCF10A as a result of the depletion of ALAD.
It was noted that the expression level of ALAD in basal-like breast cancer cell lines MDA-MB-231, MDA-MB-468, and MDA-MB-435, which ordinarily show a mesenchymal spindle-like morphology, was much lower than in the other cell lines. In addition, the mesenchymal morphology of MDA-MB-231 cells was transformed to epithelial-like by ALAD overexpression. It was supposed that ALAD would be involved in the EMT process. Cells that undergo EMT often lose the epithelial phenotype with specific markers (e.g. E-cadherin) and achieve the mesenchymal phenotype with other markers (e.g. N-cadherin, vimentin). Transforming growth factor-β signaling is thought to be the chief inducer of EMT as it plays dual roles in tumorigenesis.24, 25 Indeed, TGF-β acts as tumor suppressor during the early stage of tumorigenesis, but ultimately promotes cancer progression.26, 27 The TGF-β signaling pathway and various transcription factors, such as the Snail family, Twist, and ZEB respond to this signal and function as master controllers of the EMT program.28 It was shown in our study that ALAD was capable of inhibiting the TGF-β-induced EMT process. Cells were observed to become resistant against TGF-β stimulation as a result of ALAD overexpression.
It is understood that EMT progress is essentially acquired for most cancer cells to obtain the ability of migration and invasion, which ultimately leads to metastasis. Inhibition of TGF-β-induced EMT by ALAD could reduce the cells’ invasive capacity, which is consistent with our previous observations. It was also proved by clinical cases and the GOBO database that disease-free survival of patients with high ALAD expression is significantly longer than in other groups.
In conclusion, our work revealed the relation between ALAD and breast cancer progress. We showed that ALAD could inhibit the TGF-β-induced EMT process and reduce the invasive capacity of cancer cells. These findings suggested that ALAD might be a potential suppressor of carcinogenesis. The molecular mechanism of its detailed role in cancer progress is still unclear, and its significance in the clinic should be established by further research.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant Nos. 81372843, 81472472, and 81502518), the National Science and Technology Support Program (Grant No. 2015BAI12B15), the National High Technology Research and Development Program of China (Grant No. 2012AA021003), and the Anticancer Key Technologies R & D Program of Tianjin (Grant No. 12ZCDZSY16200).
Disclosure Statement
The authors have no conflict of interest.




