Prognostic significance of CpG island methylator phenotype in surgically resected small cell lung carcinoma
Funding Information:
The Ministry of Education, Culture, Sports, Science and Technology, Japan and grants from the Japan Society for the Promotion of Science (including grant no. 24221011), the Ministry of Health, Labour and Welfare, the Foundation for Promotion of Cancer Research in Japan, the Princess Takamatsu Cancer Research Fund, Smoking Research Foundation, Dai-ichi Sankyo Co Ltd and the NEDO project (Technology Development for Drug Discovery Platform Based on the Epigenetic Mechanism).
Abstract
Methylation is closely involved in the development of various carcinomas. However, few datasets are available for small cell lung cancer (SCLC) due to the scarcity of fresh tumor samples. The aim of the present study is to clarify relationships between clinicopathological features and results of the comprehensive genome-wide methylation profile of SCLC. We investigated the genome-wide DNA methylation status of 28 tumor and 13 normal lung tissues, and gene expression profiling of 25 SCLC tissues. Following unsupervised hierarchical clustering and non-negative matrix factorization, gene ontology analysis was performed. Clustering of SCLC led to the important identification of a CpG island methylator phenotype (CIMP) of the tumor, with a significantly poorer prognosis (P = 0.002). Multivariate analyses revealed that postoperative chemotherapy and non-CIMP were significantly good prognostic factors. Ontology analyses suggested that the extrinsic apoptosis pathway was suppressed, including TNFRSF1A, TNFRSF10A and TRADD in CIMP tumors. Here we revealed that CIMP was an important prognostic factor for resected SCLC. Delineation of this phenotype may also be useful for the development of novel apoptosis-related chemotherapeutic agents for treatment of the aggressive tumor.
DNA methylation-associated gene silencing is a common event in human cancers, including primary lung cancer.1 Global hypomethylation is thought to play a role in carcinogenesis of primary lung cancer by increasing chromosome instability.2, 3 However, local hypermethylation of promoter CpG islands consistently inactivates their downstream genes, including tumor-suppressor genes. The CpG island methylator phenotype (CIMP) displays characteristic alterations of promoter DNA methylation in colorectal cancer,4 glioblastoma5 and breast cancer.6 Poirier et al.7 report on genome-wide DNA methylation in SCLC, by using 34 fresh-frozen primary tumors, six distinct primary patient-derived xenografts and seven cell lines. In their study, the methylation patterns were clearly tied to each gene expression, and DNA methylation profiling successfully distinguished subtypes of primary SCLC tumors. However, they mentioned no clinical feature by subdividing SCLC tumors. Here we set out a program to establish a clinically-useful subclassification of SCLC using a CIMP status. Our ultimate goal is to clarify the clinical importance of the molecular biological classification.
Materials and Methods
Subjects and tumor samples
Between 1 July 1995 and 30 September 2009, a total of 1873 patients with primary lung cancer underwent surgical resection at the Cancer Institute Hospital, Japanese Foundation for Cancer Research (JFCR), Tokyo, including 49 (2.6%) SCLC patients. Among these cases, we excluded combined type SCLC tumors based on diagnosis by expert pathologists (NM and YI) using World Health Organization classification.8 In addition, we found that the tissue amount of 21 cases was insufficient for further molecular experiments. Subsequently, we were able to use 28 samples for methylation analysis, and 25 samples were available for gene expression analysis.
Cases with atypical histology were examined by a panel of Japanese expert lung pathologists organized by a neuroendocrine tumor study group,9 and tumors with consensus diagnosis were used. Written informed consent for medical research was obtained from all patients. Clinical and pathological data were stored in a database in accordance with hospital privacy rules. The study protocol was approved by the institutional review board of JFCR.
Illumina Infinium methylation assay and expression microarray analysis
After bisulfite conversion of genomic DNA, 28 samples were analyzed using Illumina's Infinium Human Methylation27 Beadchip Kit (WG-311-1202), which contains 27 578 CpG loci covering more than 14 830 human RefSeq genes at single-nucleotide resolution. All microarray datasets have been deposited into the NCBI GEO database (accession number GSE50412). All statistical analyses were carried out using β-values: (signal intensity of methylated probe)/(signal intensity of methylated probe + signal intensity of non-methylated probe), which were quantified using M-values that were calculated as the base 2 logarithm ratio of the intensities of the methylated and unmethylated probes.10
The RNA integrity number (RIN) index was calculated for each sample using Agilent 2100 Expert software, and only RNA samples with RIN number >4 were further processed. Finally, mRNA expression in 25 SCLC tissue samples was analyzed by gene expression microarray (SurePrint G3 Human Gene Expression Microarray Kit 8×60M; Agilent Technologies, Santa Clara, CA, USA). The datasets of mRNA expression have been deposited into the NCBI GEO database (accession number GSE62021).
Statistical analysis and validation using in-silico datasets
Unsupervised hierarchical clustering analysis was performed by using the Euclidean distance and complete linkage algorithm on Cluster 3.0, and the dendrogram and heat map were constructed using TreeView (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm). To reconfirm the results of hierarchical clustering, we performed non-negative matrix factorization (NMF),11, 12 a kind of consensus clustering, by using the GenePattern module (http://www.broadinstitute.org/cancer/software/genepattern#). Previously, using the software, we successfully obtained robust results of clustering for squamous cell carcinomas and adenocarcinomas of the lung.13, 14 Here we selected the number of clusters (k = 2) on NMF, based on the cophenetic correlation coefficients (k) given by the software. NMF has several advantages: it is more stable than the self-organizing map and hierarchical clustering, as well as numbers of clusters are given by the cophenetic correlation coefficients, which means the clustering is more objective, as detailed by Fujiwara et al.14 Categorical data were compared using the χ2-test or Fisher's exact test. A non-parametric approach (Wilcoxon rank-sum test) was used to determine probes/genes that are differentially methylated between the two groups of interest. Survival curves were calculated using the Kaplan–Meier method and survival distributions were compared with a log-rank test, using SPSS version 22 software (SPSS, Chicago, IL, USA). To validate our data, in-silico DNA methylation datasets of tumors, tumor cells and normal lung tissues reported previously were used as detailed below, whereas we could not perform survival analysis due to lack of follow-up data in the in-silico datasets. The Cox proportional hazards test was used to identify such factors that influenced the disease-free survival. Multivariate analyses were performed with a P-value of <0.10 in univariate analyses. A P-value of <0.05 was treated as significant.
Correlation analysis of downregulated genes and gene expression, and pathway analysis
Expression patterns and DNA methylation measurements of genes were linked to the unique genes following background correction and quantile normalization.15 A Spearman correlation test between methylation and expression was performed for the 25 tumor tissues following matching of the two datasets by Gene Symbol, using the criteria of delta-beta >0 and P < 0.05. Next, multiple-testing P-values were adjusted using the Benjamini and Hochberg method,16 and gene lists were constructed on the basis of genes that were observed to have a correlation coefficient (Spearman) of less than zero. Then, we selected the most differentially-methylated CpG sites with a false discovery rate (FDR)-adjusted P-value cut-off of 0.05. Finally, we confirmed the statistically-significant hypermethylation by using the other dataset reported previously.17 The Infinium probes were converted by the genomic position (NCBI, v36) of probe annotation for NimbleGen tiling microarrays. Gene ontology analysis for these genes was performed using DAVID Bioinformatics Resources version 6.7 (http://david.abcc.ncifcrf.gov/tools.jsp).18
Results
Clinicopathology of patients with small cell lung cancer
Clinicopathological characteristics of 28 patients examined are summarized in Table 1. They were mostly male (20/28, 71%) and 24 (86%) underwent lobectomy or more extensive surgery. Of the 28 patients, 12 (43%) were node-negative, 11 of whom had p-stage I tumors, and overall 5-year survival was 69.4%. Of the 28 cases enrolled, both tumor and normal lung tissues were available for 13 cases. So, we used the 13 paired tissues for comparison studies between tumor and normal tissues. We focused on surgical SCLC cases in this study because such cases may include early-stage as well as advanced-stage cases. Tumors with advanced stages are usually found in patients whose tumors are unresectable and only biopsy specimens are available.
| Characteristic | Cluster 1 | Cluster 2 | P-value |
|---|---|---|---|
| Number (n = 28) | 9 | 19 | |
| Age, years | 68.6 ± 5.9 | 67.0 ± 6.8 | 0.547 |
| Gender, male | 4 (44%) | 16 (84%) | 0.030 |
| Smoking (pack years) | 33.8 ± 22.4 | 50.8 ± 22.2 | 0.069 |
| Chemotherapy | |||
| Preoperative | 4 (44%) | 5 (26%) | 0.337 |
| Postoperative | 6 (67%) | 16 (84%) | 0.291 |
| Surgical procedure | |||
| Limited surgery | 1 (11%) | 3 (16%) | 0.741 |
| pT factor (T1/T2/T3/T4) | 3/4/1/1 | 13/5/1/0 | 0.227 |
| pN factor (N0/N1/N2) | 3/3/3 | 9/6/4 | 0.721 |
| IHC stain (positive) | |||
| (1) Each marker | |||
| Chromogranin A | 8 (89%) | 9 (53%) | 0.067 |
| Synaptophysin | 8 (89%) | 10 (59%) | 0.114 |
| CD56/NCAM | 9 (100%) | 14 (82%) | 0.1802 |
| (2) All three markers | |||
| All positive | 7 (78%) | 7 (41%) | 0.075 |
| One or two positive | 2 (22%) | 8 (80%) | 0.216 |
| ly (positive/negative/NA) | 9/0/0 | 11/7/1 | 0.030 |
| v (positive/negative/NA) | 8/1/0 | 15/3/1 | 0.702 |
| p (positive/negative/NA) | 5/4/0 | 5/14/0 | 0.132 |
| pm (positive/negative/NA) | 2/7/0 | 0/19/0 | 0.033 |
- Data of age and smoking: mean ± SD. IHC, immunohistochemical.
Comparisons of DNA methylation patterns between tumors and normal tissues, using both our own data and in-silico data for validation analysis
After genome-wide DNA methylation sequencing of 28 tumor tissues and 13 normal lung tissues, we found 2397 that had an SD of mean β-value that met the >0.2 threshold within the 13 tumor sample set. To identify differences of global methylation patterns between cancerous and normal tissues, β-values of the 13 paired tumor and normal tissues were analyzed using the hierarchical clustering and the NMF. Among of 2397 sites with an SD of mean β-value larger than 0.2 within tumor samples, 147 candidate sites were selected and analyzed by excluding the sites with no important statistical differences between tumor and normal tissues (Mann–Whitney U-test, P < 0.05), and values ([β-value of tumor] – [β-value of normal tissue]) smaller than 0.01 in 2 of 13 tumors (Table S1). As shown in Figure 1(a,b), the results were very similar for the two clustering methods, and tumor and normal samples were clearly clustered, implying that the clusters were quite robust. Gene ontology analyses of the 2397 loci implicated three main pathways involved in the etiology of these cancers: neuroactive ligand-receptor interaction, calcium signaling pathway and gap junction (Table S2).
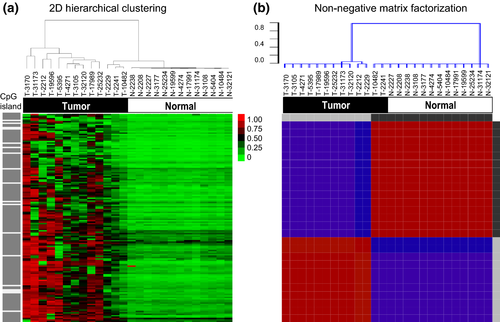
The validity of DNA methylation patterns between cancerous and normal tissues in independent cohorts obtained from the GEO database
Using the 147 genes mentioned above, we verified this gene set to be discriminable between malignant and benign lung tissues. The DNA methylation datasets of tumors and normal lung tissues reported previously17 were used as an independent validation set: GSE35341 downloaded from NCBI GEO data repository http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE35341. We employed 18 SCLC tumor tissues (T1–T18) and five normal lung tissues (N1–N5) from the series to confirm our results. To apply the probes of the Illumina Infinium HumanMethylation27 microarray to that of the NimbleGen tiling arrays, all sequences of each probe set were compared and adjusted approximately. According to the result of clustering analysis in GSE35341, the 147 probes were able to distinguish cancerous from non-cancerous specimens sufficiently: 17 out of 18 SCLC were correctly classified as “tumor” and 4 out of 5 normal lung tissues were correctly classified as “healthy” (Fig. 2).

Subclassification of small cell lung cancer by methylation patterns
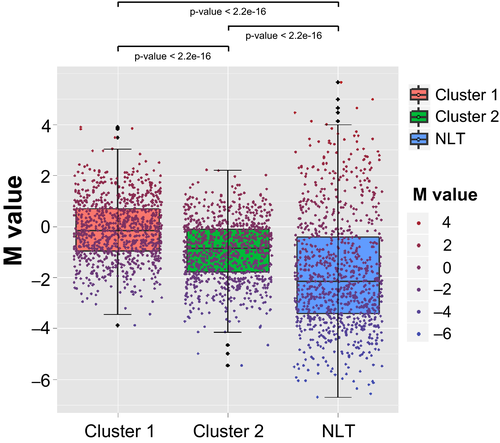
Next, to identify SCLC subgroups, we used the 1741 probes with an SD of mean β-value in tumor tissues >0.2 on each site and performed both the unsupervised hierarchical clustering (Fig. 3a) and the NMF (Fig. 3b). We identified two clusters with different methylation levels: Cluster 1 (n = 9) and Cluster 2 (n = 19). As shown in Figure 3(a,b), the two clusters created by both the methods were exactly the same, implying that the clusters were very robust. Cluster 1 tumors were identified as SCLC CIMP and Cluster 2 was non-CIMP, because the CpG islands of Cluster 1 tumors were significantly hypermethylated as compared with Cluster 2 tumors (Fig. 4).
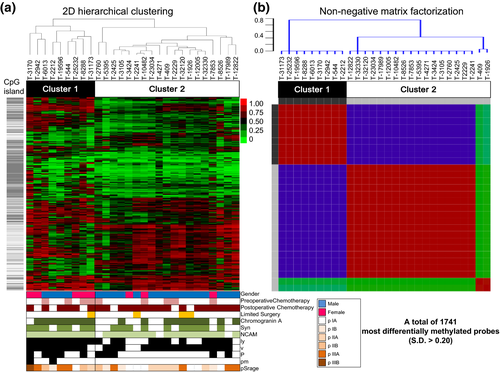
Clinical, pathological and immunohistochemical data of SCLC in the two subgroups are summarized in Table 1. Comparison of the baseline characteristics between these two groups showed significant differences in gender, ly and pm factors. Furthermore, tumors of Cluster 2 tended to have a less neuroendocrine nature, such as chromogranin A (89% Cluster 1 vs 53% Cluster 2), synaptophysin (89% vs 59%) and all markers (78% vs 41%), although the tendencies were not statistically significant, which agrees with our previous study results based on neuroendocrine marker expression.19
Survival analysis
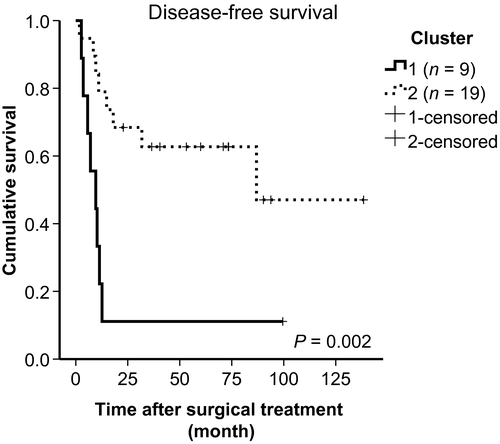
During the median follow-up period of 37.4 months, 16 patients suffered cancer relapse: 8 (8/9, 89%) in Cluster 1 and 8 (8/19, 42%) in Cluster 2. The 5-year disease-free survival (DFS) rate for the entire group was 46.2%, and the 5-year DFS of Cluster 1 (11.1%) was much lower than that of Cluster 2 (62.7%). These differences are highly significant (Fig. 5, P = 0.002).
Univariate and multivariate analyses for prognosis
Next we performed multivariate analyses for prognosis, by using variables with a value of P < 0.10 in the univariate analyses (Table 2). Postoperative chemotherapy was a significant good prognostic factor, and being a Cluster 1 patient (SCLC CIMP) was a poor prognostic factor (Table 3).
| Variable | Hazard ratio | 95% CI | P-value |
|---|---|---|---|
| Age, years | 0.972 | (0.888, 1.064) | 0.539 |
| Gender, male | 0.641 | (0.221, 1.862) | 0.414 |
| Smoking | 0.979 | (0.956, 1.002) | 0.079 |
| Preoperative chemotherapy | 2.593 | (0.926, 7.264) | 0.070 |
| Postoperative chemotherapy | 0.198 | (0.069, 0.565) | 0.002 |
| Limited surgery | 1.377 | (0.387, 4.897) | 0.621 |
| pT factor | 1.853 | (1.025, 3.350) | 0.041 |
| pN factor | 1.518 | (0.819, 2.814) | 0.185 |
| NE marker immunostaining | |||
| Chromogranin A | 3.011 | (0.853, 10.62) | 0.087 |
| Synaptophysin | 2.514 | (0.712, 8.875) | 0.152 |
| NCAMa | 27.68 | (0.099, 7707) | 0.248 |
| All positive | 2.111 | (0.732, 6.095) | 0.167 |
| pl 1–3 | 1.347 | (0.488, 3.723) | 0.565 |
| pm (+) | 4.858 | (0.970, 24.34) | 0.055 |
| v (+) | 3.062 | (0.402, 23.31) | 0.280 |
| ly (+) | 2.041 | (0.580, 7.186) | 0.267 |
| Cluster 1 (SCLC CIMP) | 4.399 | (1.578, 12.27) | 0.005 |
- a For NCAM, an exact CI was not able to be calculated because of no negative cases in Cluster 1. CI, confidence interval; CIMP, CpG island methylator phenotype; NE marker, neuroendocrine marker including chromogranin A, synaptophysin and NCAM; SCLC, small cell lung cancer.
| Variable | Hazard ratio | 95% CI | P-value |
|---|---|---|---|
| Postoperative chemotherapy | 0.179 | (0.057, 0.557) | 0.003 |
| Cluster 1 (SCLC CIMP) | 4.708 | (1.553, 14.27) | 0.006 |
- CI, confidence interval; CIMP, CpG island methylator phenotype; SCLC, small cell lung cancer.
Correlation of gene expression with their methylation patterns, and pathway analysis
Differentially expressed genes were identified by comparing Cluster 1 (n = 9) and Cluster 2 (n = 16). To validate if methylated genes were actually downregulated or not, we examined correlation of methylation with expression of genes by using the 1741 methylation data with the gene expression datasets of 58 724 probes. 1530 CpG sites (corresponding to 1220 genes) were identified, and 46 genes of them (corresponding to 55 CpG loci) were negatively correlated with the FDR-adjusted P-value < 0.05 (Table S3). Furthermore, tumor necrosis factors (TNFRSF10A and TNFSF8) and apoptosis factors (TANK and TRADD) were closely related to the poor prognosis of Cluster 1 patients.
The functional enrichment analysis for these genes revealed three biological pathways. Two of them were expressed with significant differences and are listed in Table S4, termed hsa04210 (apoptosis) and hsa05223 (non-small cell lung cancer) (P = 0.0067 and 0.00243, respectively) in KEGG pathways. This implies that apoptotic activities and characteristics related to non-small cell carcinoma (NSCLC) are reduced in Cluster 1 tumors by hypermethylation.
Discussion
High throughput methylation platforms enable us to reveal extensive methylation profiling of a large number of genes for a variety of human cancers, including SCLC. In this study, we found that the 147 probes could be a molecular classifier between cancerous and non-cancerous specimens (Fig. 1). This probe list was validated by using Kalari's dataset (Fig. 2).17 In addition, our functional annotation analysis demonstrated close relevance between SCLC tumors and the neuroactive ligand-receptor interaction pathway. Considering both the reports of Kalari et al. and the present report, loss of proper neuronal differentiation may be involved in the progression process of carcinoma cells to a more highly malignant stage (i.e. SCLC).
Previously, we identified two subgroups with different clinical outcomes by gene expression profiling using 38 surgically resected high-grade neuroendocrine tumors, including 15 pure type SCLC tumors19, and confirmed by immunohistochemistry of neuroendocrine markers.20 Of the two subgroups, one subset (termed non-HGNT2) showed significantly worse prognosis compared with another (5-year survival 12% vs 83%; P = 0.0094). Here, by applying DNA methylation profiling techniques, we obtained two clusters with different methylation patterns and different prognosis: Cluster 1 with global high CIMP and poor prognosis (SCLC CIMP) and Cluster 2 with low CIMP and better prognosis (non-CIMP). Poirier et al.7 also observed distinct subtypes of SCLC by using a DNA methylation profiling technique. In their study, the three clusters (termed M1, M2 and SQ-P) were identified with each different methylation pattern as well as with distinct gene expression. They confirmed that two subtypes (M1 and M2) were significantly more frequently methylated compared with the SQ-P. They discussed the relationship between the methylation status and the biological aggressiveness of tumor itself, but did not show any clinical data linked with the results. We successfully showed that the increasing methylation level was related to the poor prognosis. This suggests the possibility that it could be useful for surgical indication and for determining adjuvant therapy.
A number of genes with higher methylation levels and downregulated gene expression proved to be involved in the apoptosis and NSCLC pathways. To our knowledge, there are few reports describing that these pathways might be strongly related with the therapeutic results of SCLC. The apoptotic pathway plays an essential role in the development and maintenance of tissue homeostasis,21 and has an important role in carcinogenesis, cancer progression and resistance to anti-cancer agents.22 It has been reported that the apoptosis pathway can be effectively inactivated in SCLC cells.23, 24 The authors of these reports found resistance to FasL and TRAIL-induced death of SCLC cells, and identified silencing of Fas, TRAIL-R1 and caspase-8 expression caused by DNA methylation. In the present study, we found that SCLC CIMP had a particularly worse outcome compared with that of non-CIMP, and showed markedly lower expression of TNFRSF1A, TNFRSF10A and TRADD. Among them, TNFRSF10A was the most frequently hypermethylated and downregulated gene, inactivation of which was also reported previously in osteosarcomas,25 gastric carcinomas26 and glioblastoma multiforme.27 These results strongly suggested an important prognostic role of epigenetic suppression of TRADD.
This is the first paper that describes the 5-year DFS rates of distinct molecular SCLC subgroups (SCLC CIMP and non-CIMP). However, there were some limitations of our study: (i) the small number of cases; (ii) all samples were surgically resected; (iii) there was no validation set; and (iv) the influence of preoperative chemotherapy. Because the mainstream treatment for SCLC patients is chemo-radiotherapy or chemotherapy alone, surgical treatment is seldom undertaked. Therefore, we rarely obtain sufficient fresh materials for research. Even when samples can be obtained, preoperative chemotherapy makes analyses difficult. In this study, 9 (32%) of 28 SCLC patients underwent chemotherapy before surgery. To maintain the statistical power, we could not avoid excluding these cases. Similarly, we could not design an additional validation test. In future we need further studies with increased numbers of cases, including extended and pretreated tumors.
In summary, we revealed that SCLC CIMP could be an important prognostic indicator after surgical treatment. This may be a useful resource for surgical indication for SCLC patients, especially with cT1N0M0 stage tumors, and may help in the development of novel chemotherapeutic agents, including demethylating agents.
Acknowledgments
We thank Ms Kimie Nomura, Ms Hiroko Nagano and Ms Hiroko Meguro for excellent technical assistance. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan and grants from the Japan Society for the Promotion of Science (including grant no. 24221011), the Ministry of Health, Labour and Welfare, the Foundation for Promotion of Cancer Research in Japan, the Princess Takamatsu Cancer Research Fund, Smoking Research Foundation, Dai-ichi Sankyo Co Ltd and the NEDO project (Technology Development for Drug Discovery Platform Based on the Epigenetic Mechanism).
Disclosure Statement
Yuichi Ishikawa received funding from Dai-ichi Sankyo, Chugai Pharmaceutical and Sony, and is a consultant of Fujirebio. The other authors have no conflict of interest to declare.




