Intracellular signals of lung cancer cells as possible therapeutic targets
Funding Information:
Kansai Medical University; Grant-in-Aid for Scientific Research; Funding Program for Next Generation World-Leading Researchers; The Mochida Memorial Foundation; The Naito Memorial Foundation; The Cell Science Research Foundation; The Uehara Memorial Foundation; The Mitsubishi Foundation; The Yasuda Memorial Foundation; The Takeda Science Foundation.
Abstract
In recent years, several molecularly targeted therapies have been developed as part of lung cancer treatment; they have produced dramatically good results. However, among the many oncogenes that have been identified to be involved in the development of lung cancers, a number of oncogenes are not covered by these advanced therapies. For the treatment of lung cancers, which is a group of heterogeneous diseases, persistent effort in developing individual therapies based on the respective causal genes is important. In addition, for the development of a novel therapy, identification of the lung epithelial stem cells and the origin cells of lung cancer, and understanding about candidate cancer stem cells in lung cancer tissues, their intracellular signaling pathways, and the mechanism of dysregulation of the pathways in cancer cells are extremely important. However, the development of drug resistance by cancer cells, despite the use of molecularly targeted drugs for the causal genes, thus obstructing treatment, is a well-known phenomenon. In this article, we discuss major causal genes of lung cancers and intracellular signaling pathways involving those genes, and review studies on origin and stem cells of lung cancers, as well as the possibility of developing molecularly targeted therapies based on these studies.
Lung cancers are malignant tumors with poor prognoses. They rank as the top cause of cancer-related deaths in the world and in Japan. The development of advanced therapies for lung cancers, including molecularly targeted therapies, is an extremely important subject. For this purpose, understanding intracellular abnormal signaling caused by causal genes of lung cancers is necessary. In addition, identifying origin cells of lung cancer and candidate lung cancer stem cells, which are predicted to exist in lung tumor tissues, and understanding the difference in tissue maintenance mechanisms between the candidate cancer stem cells and their normal counterparts are important.
Lung cancers have three main histological types, namely adenocarcinoma, squamous cell carcinoma, and SCLC. Adenocarcinoma and squamous cell carcinoma are also collectively called NSCLC. With regard to the origin cells of these three types of lung cancers, adenocarcinoma has been reported to develop from AT II cells, Clara cells, and BASCs;1-3 squamous cell carcinoma, from basal cells;4 and SCLC, from PNECs.5 However, the forced expression of Ras in PNECs, which is believed to be an origin cell of SCLC, leads to the development of adenocarcinoma.6 Furthermore, induction of p53(−)–Rb(−) in AT II cells leads to the development of SCLC, which is believed to be an origin cell of adenocarcinoma,7 indicating that, in some cases, the histological types of lung cancers are determined by causal genes rather than by origin cells. All of these origin cells of cancer are proposed to be the cells that play a central role in maintenance/regeneration of the pulmonary epithelium as stem cells/progenitors in normal lung epithelial tissues (Fig. 1). By contrast, terminally differentiated cells such as AT I and ciliated cells are currently not considered as origin cells of cancer. In malignant tumors derived from lung stem/progenitor cells such as AT II cells, Clara cells, BASCs, and PNECs, intracellular signaling, which is important for the maintenance of normal cells, should also be used for promoting survival in cancer cells.
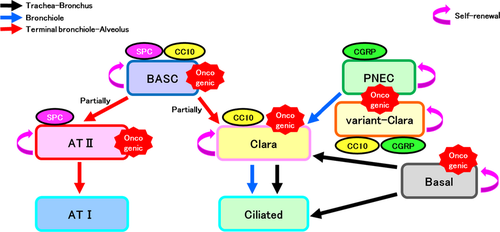
A cancer stem cell model was proposed. Malignant tumors, including lung cancers, are heterogeneous cell groups containing many cell types. Cancer stem cells present in tumors in a very small number, have long lives, and supply other more-differentiated tumor cells. The differentiated cancer cells have only limited life spans and withdraw from tumor tissues in a short period. These routinely replace most cells in tumor tissues. In this model, cancer stem cells are supposed to be an important therapeutic target because the removal of these stem cells allows destruction of the whole tumor. However, the existence of cancer stem cells has not been proven experimentally in many organ-derived malignant tumors. To accurately prove the existence of cancer stem cells in tumors, lineage tracing experiments are very important, as described later. In addition, cancer stem cell-targeted therapy has not been realized in clinical practice. In cases of malignant tumors derived from other organs, such as a hematological malignancy, a model has been proposed in which a cancer stem cell enters a niche of a normal long-term stem cell and the resting phase of the cell cycle (G0 phase) to gain resistance to anticancer agents and radiotherapy. This model is actively studied from the viewpoint of overcoming resistance to therapies. In addition, in lung cancers, cancer stem cells with resistance to lung cancer treatment through a similar mechanism may exist. However, as addressed later in this article, only limited information is currently available regarding pulmonary stem cells, lung cancer stem cells, and their niches.
In this article, signals and transcription factors considered useful as future targets of therapies that target cancer stem cells are reviewed from the perspective of lung development and lung epithelial regeneration. Involvement of epigenetics and microRNAs in the onset and survival signals of lung cancer are also mentioned. Finally, we also introduce our studies in which we used multicolor lineage tracing for the identification of origin cells of lung cancers and lung cancer stem cells.
Tyrosine Kinase Family
Major causal genes known to induce NSCLC include EGFR and KRAS. More than 90% of activating mutations of the EGFR gene are L858R mutations and exon 19 deletions. An additional T790M mutation in exon 20 to these activating mutations causes drug resistance to EGFR–TKIs, which is the most frequent mechanism known for EGFR–TKI-resistant lung tumors.8 A T790M mutation of EGFR, such as the T315I mutation of ABL in chronic myeloid leukemia patients, is considered to inhibit binding of TKI to the ATP binding site of the kinase domain. Other mechanisms of EGFR–TKI resistance include amplification of c-MET and the resulting activation of the PI3K pathway,9 and overexpression of HGF, which causes c-MET activation.10
In 2007, the EML4–ALK transforming fusion gene was identified in NSCLC patients.11 Treatment with ALK–TKI has been given to NSCLC patients with the EML4–ALK fusion gene, and this has achieved dramatically good results. However, the drug resistance mutation of L1196M of the ALK gene, which corresponds to T790M of EGFR and T315I of ABL mentioned earlier, was previously reported.12 Moreover, transforming fusion genes involving ROS1 or RET receptor tyrosine kinases have also been identified, and development of specific TKIs for these fusion genes is underway. Although these TKIs are currently among the most successful molecularly targeted therapies for lung cancers, other categories of signaling molecules could possibly be targets for next-generation molecularly targeted therapies. Therefore, they will be discussed in the following sections.
Notch Pathway
Notch signaling is involved in lung development, bronchiolar epithelial regeneration, and lung cancer development (see Fig. 2 for an overview). Notch signaling is reported to be activated in NSCLC, and it facilitates self-renewal and EMT of cancer stem cells.13, 14 In mammals, four Notch types (Notch1–4) are known. Notch113, 15 and Notch3 signaling16, 17 are reported to be activated in NSCLC. A synergistic effect of Notch and c-Myc has also been reported.18 c-Myc, as well as Hes and Hey, among others, is a target gene of Notch signaling. The fact that c-Myc is also involved in signaling through Sox2 or STAT3 suggests the presence of cross-talk among various signaling pathways through c-Myc. Reported Notch activation mechanisms observed in NSCLC include deletion of NUMB expression and gain-of-function mutation of the NOTCH1 gene.19 In addition, regarding Notch3 signaling, knockdown of NOTCH3 leads to downregulation of the anti-apoptotic genes BCL-2/BCL-XL and upregulation of the apoptosis-promoting genes BAX/BIM/BAD.20, 21 BCL-XL is highly expressed in NSCLC,22 whereas BIM is downregulated in EGFR–TKI-resistant lung cancer cells.23 In contrast to Notch1 and Notch3, Notch2 is reported to function in a tumor-suppressive manner in NSCLC.24 Meanwhile, as previously mentioned, Notch signaling plays a role in tumor proliferation in NSCLC in most cases, whereas Notch expression functions in a tumor-suppressive manner in SCLC, which may suggest that Notch signaling plays suppressive roles in SCLC stem cells. Activation of Notch1 or Notch2 signaling in SCLC, in which the expression level of NOTCH is low, leads to inhibition of tumor proliferation.25, 26 Pulmonary neuroendocrine cells, which are believed to be the origin cells of SCLC, are progenitor cells of Clara cells. In lung development, Notch signaling regulates the differentiation of PNECs into Clara cells.27, 28 In the mouse model of regeneration after naphthalene-induced bronchiolar epithelial injury, Notch1 signaling is important for the regeneration of Clara cells. After Clara cells have been completely removed through the naphthalene injury, PNECs have proliferated transiently, differentiating into Clara cells and then into ciliated cells.29 Given these events, the tumor-suppressive effect of Notch signaling in SCLC may lie in the induction of differentiation of SCLC stem cells with characteristics similar to those of PNECs. In the alveolar region, Notch signaling is suggested to regulate development/differentiation and regeneration.30, 31 Moreover, Notch signaling is reported to suppress a phosphatase (dual-specificity phosphatase 1) and activate EGFR/ERK1/2 signaling.24, 32

Sox2
The SOX2 gene is highly expressed in all of the three histological types of lung cancer, namely adenocarcinoma, squamous cell carcinoma, and SCLC.33 SOX2 is involved in lung development and bronchial epithelium regeneration, and suppression of SOX2 leads to Hedgehog signal reduction. Moreover, SOX2 expression is detected in the airway epithelium, but not in the alveolar region, in the process of lung development.34 In adenocarcinoma, inhibition of SOX2 expression induces apoptosis of tumor cells35 and downregulation of WNT1/2, NOTCH1, and c-MYC; it also decreases the number of SP cells.33 Singh et al.36 reported that inhibition of EGFR/AKT signaling in SP cells of NSCLC induces suppression of SOX2 expression, which in turn suppresses self-renewal of SP cells, indicating the involvement of EGFR signaling and SOX2 expression in stem cell maintenance. Chou et al.37 reported a positive feedback loop via c-MYC between SOX2 and EGFR, in which c-MYC binds to the SOX2 promoter region after EGFR stimulation to upregulate SOX2. In addition, SOX2 suppresses apoptosis by upregulating EGFR and BCL-XL expressions by binding to their promoters. Thus, many reports stated that Sox2 plays a facilitative role in tumor development/proliferation of NSCLC, whereas Xu et al. reported that Sox2 acts for suppression of NSCLC. They reported that Sox2 suppresses the expressions of Notch1, Notch2, and Hes1 by binding to their promoters, although Notch signaling is required for Kras mutation-induced adenocarcinogenesis.38
As one of the four Yamanaka factors for induced pluripotent stem cell induction, SOX2 is known to be important for normal and cancer stem cells in some other tissues. As discussed, SOX2 plays critical roles in the self-renewal of NSCLC stem cells.33, 35, 36, 39 SOX2 is also amplified in ~27% of SCLC and contributes to SCLC proliferation.40 In the trachea, where basal, Clara, and ciliated cells exist, SOX2 deletion reduces the numbers of each cell and epithelial cell density, which could suggest involvement of SOX2 in maintenance/regeneration of the trachea epithelium.34 The role of SOX2 in alveolar maintenance/regeneration has not been elucidated.
WNT Pathway
The canonical Wnt signaling pathway (Fig. 3), in which β-catenin is involved, as in many other tissues, is important in lung cancer proliferation, lung development, and regeneration after injury. WNT signaling is reported to be activated in NSCLC stem cells.41 In NSCLC, activation of WNT signaling induces tumor proliferation and acquisition of metastatic potential.41, 42 In addition, inhibition of WNT2 signaling leads to downregulation of Survivin, an anti-apoptotic gene; this consequently induces apoptosis.43 In the lung adenocarcinoma mouse model produced by Kras mutation and Wnt signaling activation, enhanced proliferation and EMT of lung cancer cells, as well as downregulation of Sox2 and upregulation of Sox9 and Gata6, were observed, in comparison with the lung adenocarcinoma mouse model produced by Kras mutation only.44 In lung development, Wnt signaling is thought to negatively regulate Sox2;45 therefore, it may function similarly in NSCLC.
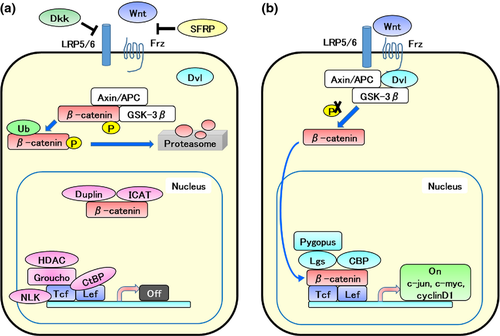
Stabilization of β-catenin blocks Clara cell differentiation to ciliated cells and increases the number of naphthalene-resistant progenitor cells.46 Naphthalene resistance is the characteristic of variant Clara cells. In addition, deletion of β-catenin in basal cells suppresses proliferation and leads to apoptosis.47 Moreover, autocrine secretion of insulin-like growth factor-I from AT II promotes differentiation of AT II to AT I-like cells through activation of the non-canonical Wnt pathway regulated by Wnt5a.48
STAT Pathway
The JAK/STAT signaling, as well as the RAS/ERK1/2 and PI3K/AKT signaling, is a representative intracellular signaling pathway downstream of receptor tyrosine kinases and cytokine receptors. In NSCLC with EGFR mutation, activation of STAT and/or AKT is involved in tumor proliferation.49 According to Zhao et al.,50 STAT3- and STAT3-induced c-MYC facilitate SOX2 expression, and SOX2 possesses a positive feedback action to STAT3 and c-MYC. Interleukin-6 facilitates proliferation of Kras-mutated NSCLC.51 The JAK/STAT signaling pathway, a signaling downstream of the IL-6 receptor, may be involved in this effect. In addition, as IL-6 is reported to contribute to survival and maintenance of cancer stem cells through NOTCH3 signaling in breast cancer,52 Notch signaling may also be involved in the mechanism of proliferation promotion by IL-6 in NSCLC. Although the involvement of JAK/STAT signaling in normal lung stem cell is not well known, STAT3, by interacting with SOX2, is reported to contribute to the self-renewal of lung cancer stem cells.50
E-cadherin
E-cadherin is used as an indicator of EMT in many cancers and tumors. Because deletion of E-cadherin leads to nuclear translocation of β-catenin, which binds to the intracellular domain of E-cadherin, the deletion activates the Wnt/β-catenin signaling. In the process of alveolar epithelial regeneration, both E-cadherin(+) and (−) AT II cells are observed. Among them, E-cadherin(−) AT II cells are suggested to function as stem cells of the alveolar epithelium because a correlation between an increase of telomerase activity and high proliferation potential are observed in E-cadherin(−) AT II cells.53 Knockdown of E-cadherin expression in AT II cells leads to development of hyperplasia,54 and suppression of E-cadherin expression in Clara cells leads to the appearance of ectopic BASCs and an increase in the number of variant Clara cells.55 From these findings, downregulation of E-cadherin is likely to upregulate self-renewal of lung stem cells.
Epigenetic Modification and Lung Cancer
DNA methylation is abnormally enhanced in pulmonary precancerous lesions and lung cancer cells. Methylation of gene regulatory regions is important as an inactivation mechanism of tumor suppressor genes because the methylation strongly suppresses the expression of downstream genes. Meanwhile, methylation of coding regions may promote oncogene expression. The post-translational modification of the N-terminal region of histone, including acetylation, methylation, ubiquitination, phosphorylation, and sumoylation, regulates DNA transcription and replication/repair. As it is said in the cases of Helicobacter pylori infection in stomach cancer, and hepatitis B and C virus infections in liver cancer, chronic inflammation might lead to cancer development through genomic DNA methylation. In fact, in some cases of inflammatory diseases of the lung, including interstitial pneumonia, obsolete pulmonary tuberculosis, non-tuberculosis mycobacteriosis, and chronic pleural empyema, lung cancer complications often occur. In those cases, DNA methylation of pulmonary epithelial cells might be one cause of lung cancer development. A report also stated that DNA methylation occurs prior to gene mutation and/or deletion, and accumulation of DNA methylation leads to cancer formation.56 In stage I lung squamous cell carcinoma, several hundreds of CpG islands are hypermethylated.57 In NSCLC, high methylation of P16, RASSF1, APC, and H-cadherin is observed.58 In addition, detection of methylation of P16 or a DNA repair enzyme, the MGMT gene in sputum cells is reported to be useful for early detection of NSCLC.59 In addition, DNMT1 is upregulated in precancerous lesions and cancer tissues. In lung cancer, DKK3, a secreted Frizzled-related protein (SFRP1, SFRP2, and SFRP5), which is a negative regulator of the WNT pathway, is hypermethylated and suppressed.60 Among molecules in the Notch pathway, NOTCH3, JAG1, HES2, HES4, and HES5 have been reported to be hypermethylated in leukemia,61 but the epigenetic status in the Notch pathway in lung cancer has not been revealed.
Histone modification has many mechanisms including acetylation and methylation, which are highly important in transcription regulation. Regarding histone methylation, facilitation or suppression of transcription depends on the type of targeted amino acid residue and its location in the histone N-terminal. In particular, methylation of Lysin27 of Histone H3 (H3K27) is important as an inactivation mechanism of tumor suppressor genes. In fact, the EZH2 gene, an H3K27 methyltransferase, is upregulated in breast and prostate cancers.62 The same mechanism might function in lung carcinomas.
Although involvement of epigenetics in lung cancer has not been well investigated, WNT signaling could be activated by methylation of APC,58 SFRP1, SFRP2, SFRP5, and DKK360 in a subset of lung cancer. Wnt signaling is involved in lung cancer stem cells and lung maintenance/regeneration;41, 46-48 therefore, epigenetic modification in Wnt signaling plays some roles in lung cancer and normal lung stem cells.
MicroRNA and Lung Cancer
MicroRNAs are non-coding RNAs that suppress gene expressions by inhibiting translation and/or stability of target gene mRNAs. In lung cancers, miR-29 targeting DNMT3A/3B is downregulated,63 as well as let-7 targeting KRAS64 and miR-487b targeting KRAS, MYC, WNT5a, BMI1, and SUZ12.65 In contrast, miR-31 targeting DKK1 is upregulated.66 Moreover, in lung cancer, miR-34, 199a*, and 7515 targeting c-Met are suppressed.67-69 Furthermore, miR-17-92 is known to be upregulated mainly in SCLC.64 In a gastric cancer cell line, miR-126 targets SOX2.70 The survival signaling, epigenetics, and miRNAs related to the development of lung cancers that were mentioned earlier are outlined in Figure 4.
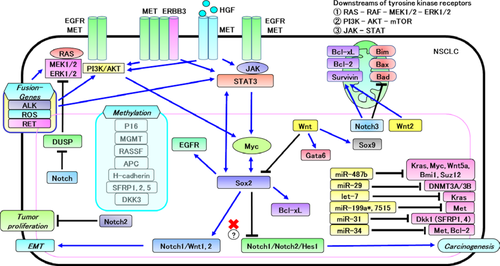
Mechanisms of Drug Resistance of Tumor Cells in Lung Cancers
A proportion (20–30%) of lung cancer patients with EGFR mutation is primarily resistant to EGFR–TKIs. In addition, in 50% of lung cancer cases with KRAS mutation, KRAS knockdown exerts no effect on tumor cell proliferation.71 These indicate that development of lung cancers, even those with a mutation of a popular lung cancer-related genes such as EGFR and KRAS, is not necessarily solely dependent on these genes, but another signal pathway is likely to function as survival signaling. Indeed, in addition to the EGFR gene mutation (T790M mutation), c-MET amplification and HGF overexpression are well-known bypass mechanisms of EGFR signals. In addition, as has been mentioned here, c-MYC is likely to play the role of survival signaling mediator in NSCLC. Meanwhile, a report stated that in SCLC, Ras mutation occurs in EMT, causing downregulation of Mycl1.72 Because, as described earlier, Notch signaling, SOX2, WNT signaling, and STAT signaling are involved in lung cancer stem cells, simultaneous inhibitions of several or all of them in addition to EGFR–TKI might bring more suppressive effects on lung cancer.13, 21, 36, 42, 49, 50
Multicolor Lineage Tracing using Rainbow Mice
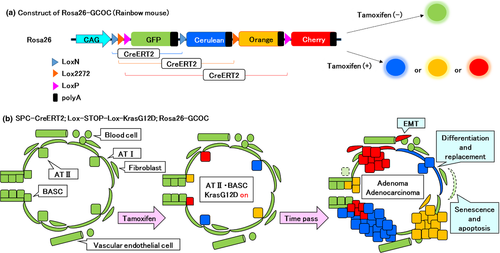
In recent years, properties of lung stem cells/progenitors and relationships between them have been gradually clarified.2, 31, 73 The lineage relationships between the various types of lung epithelial stem cells shown in Figure 1 are not yet well clarified. In addition, lineage relationships between the histological types of lung cancers mentioned in the first section of this article, and their origin cells, as well as whether cells equivalent to lung cancer stem cells exist in formed tumor tissues of lung cancer are important topics, with a view toward development of future cancer stem cell-targeted therapies. Genetic cell lineage tracing is a powerful method to analyze such lineage relationships between two different types or between more than two types of cells.74 However, the original method of lineage tracing is difficult to apply to tissues with slow cellular turnover such as lung epithelial tissues, and tissues in which many undifferentiated cells are involved in a complicated manner in the maintenance of the tissues. Therefore, we have developed multicolor lineage tracing to improve the statistical accuracy of the lineage tracing. Figure 5 shows a construct of the rainbow mice that we developed. The advantage of the method is that clonality of tissue maintenance can be visualized in situ. The method has already been used to analyze epithelial stem cells in the small intestine and germ stem cells in the testis, and to identify lingual epithelial stem cells.75-78 Using this method, Desai et al.79 showed that during fetal development, AT I and AT II arise from common progenitor cells; after birth, AT I is derived from rare AT II with a self-replication potency. These findings offer prospects of future applications of such techniques for analyses of interlineage relationships between multiple proposed stem cells in the lung epithelium, identification of origin cells of lung cancers, and analyses of lung cancer stem cells in order to answer the many questions that remain.
Conclusion
Many lung cancer-related oncogenes have been identified, and abnormalities of signaling pathways in lung cancer cells, including cancer stem cells, have been revealed. In addition, not only genomic mutation but also epigenetic abnormality has been reported as important in the onset of development of lung cancers. Chemotherapies based on histological classifications should shift to individual therapies based on causal genes and epigenetic abnormalities. Moreover, in the future, molecularly targeted therapies that are selectively effective for chemotherapy-resistant, lung cancer stem cells will be developed. From this standpoint, understanding the mechanisms of survival signals and epigenetic abnormalities related to lung cancer/stem cells, which are discussed in this review, should be extremely important.
Acknowledgments
The authors thank the members of the Department of Stem Cell Pathology, Kansai Medical University (Hirakata, Japan) for helpful discussions. We acknowledge financial support from the following sources: Kansai Medical University Study Grant to K. T. and K. K., Grant-in-Aid for Scientific Research to K. K., Funding Program for Next Generation World-Leading Researchers, The Mochida Memorial Foundation, The Naito Memorial Foundation, The Cell Science Research Foundation, The Uehara Memorial Foundation, The Mitsubishi Foundation, The Yasuda Memorial Foundation, and The Takeda Science Foundation to H.U.
Disclosure Statement
The authors have no conflict of interest.
Abbreviations
-
- ALK
-
- anaplastic lymphoma kinase
-
- AT (I/II)
-
- alveolar type (I/II)
-
- BASC
-
- bronchioalveolar stem cell
-
- EGFR
-
- epithelial growth factor receptor
-
- EMT
-
- epithelial–mesenchymal transition
-
- IL
-
- interleukin
-
- NSCLC
-
- non-small-cell lung cancer
-
- PI3K
-
- phosphoinositide-3-kinase
-
- PNEC
-
- pulmonary neuroendocrine cell
-
- SCLC
-
- small-cell lung cancer
-
- SP
-
- side-population
-
- STAT
-
- signal transducer and activator of transcription
-
- TKI
-
- tyrosine kinase inhibitor




