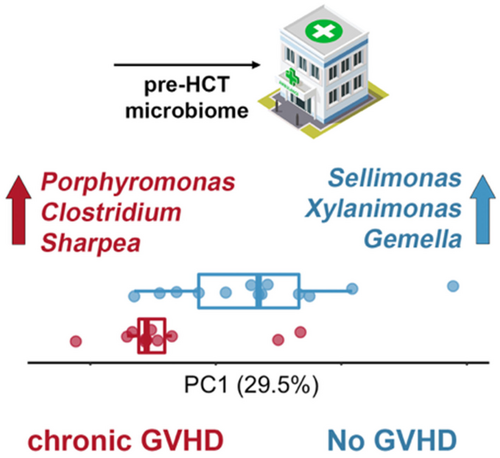
Pre-transplant and longitudinal changes in faecal microbiome characteristics are associated with subsequent development of chronic graft-versus-host disease
Summary
The role of the gastrointestinal microbiome in predisposing to chronic graft-versus-host disease (cGVHD), an immune-mediated haematopoietic cell transplant (HCT) complication, is not well defined. We examined the relationship of the host faecal microbiome with subsequent cGVHD development by analysing baseline stool samples as well as post-HCT changes in microbiome composition and metabolite pathway analyses. We analysed pre-transplant baseline samples from 11 patients who subsequently developed cGVHD compared to 13 controls who did not develop acute GVHD or cGVHD at any time. We found a significant differential abundance of multiple taxa at baseline between cGVHD versus controls, including the Actinobacteria phylum and Clostridium genus. A subgroup analysis of longitudinal samples within each patient revealed a greater loss of alpha diversity from baseline to post-engraftment in patients who subsequently developed cGVHD. Metabolic pathways analysis revealed that two pathways associated with short-chain fatty acid metabolism were enriched in cGVHD patient microbiomes: β-oxidation and acyl-CoA synthesis, and γ-aminobutyrate shunt. In contrast, a tryptophan catabolism pathway was enriched in controls. Our findings show a distinct pattern of baseline microbiome and metabolic capacity that may play a role in modulating alloreactivity in patients developing cGVHD. These findings support the therapeutic potential of microbiome manipulation for cGVHD prevention.
Graphical Abstract
INTRODUCTION
Chronic graft-versus-host disease (cGVHD) is a multisystem syndrome involving dysregulated immunity, tissue inflammation/injury, and excessive tissue fibrosis often leading to long-term disability after allogeneic haematopoietic cell transplantation (HCT).1 While the relationship between gastrointestinal microbiota and acute GVHD (aGVHD), overall survival, infections and other post-transplant complications has been described previously,2-6 the role of the gastrointestinal microbiome in predisposing to the immune-mediated HCT complication of cGVHD is not well defined.7-10 Gut microbiome-derived metabolites, including short-chain fatty acids (SCFA), secondary bile acids, and aryl hydrocarbon receptor ligands have been shown to influence immune responses in the intestines and the gut epithelial barrier.11, 12 These changes could subsequently affect patient susceptibility to post-transplant morbidities including cGVHD.
We examined the potential relationship of the host faecal microbiome with subsequent cGVHD development by analysing baseline stool samples at the time of HCT for microbiome composition and inferred function.
METHODS
We identified cGVHD patients in our prospective study approved by the University of Minnesota Institutional Review Board for whom faecal samples were available and sequenced. We analysed pre-transplant baseline faecal samples from 11 adult patients who subsequently developed cGVHD and compared those to baseline stool samples of 13 controls who did not develop aGVHD or cGVHD at any time post-transplant. From this set of patients, we performed a subgroup analysis on post-engraftment longitudinal samples (between day 49 and 130 post-transplant) that were also available for five of the patients with cGVHD and four controls.
Inpatient faecal samples were collected on swabs or from the whole stool and frozen at −80°C until processing; outpatient (longitudinal) samples were collected in RNAlater and homogenized with glass beads, and aliquoted and frozen at −80°C upon laboratory receipt. Samples were submitted to the University of Minnesota Genomics Center (UMGC) for DNA extraction, Illumina Nextera XT (1/4 reaction) library preparation, and shotgun metagenomic sequencing on Illumina HiSeq 2500 (2 × 125 bp) or NovaSeq (2 × 150 bp) platforms. Raw forward DNA reads were processed and converted to FASTA using SHI7 v0.9.913 with a minimum quality of 34 and length of 80, after which the median sample depth was 4.68 million reads. Human-aligning reads (mean 2.6%, median 0.14% of reads, range 0.0005%–34%) were removed using SHOGUN filter,14 and the remaining reads (read depth median 4.67 million, mean 8.89 million, range 37 416–34 310 916) aligned using the optimal alignment tool BURST at 98% identity as previously described.15-17 Features (references) with <1000 counts or <5% prevalence were removed. Alpha diversity metrics were calculated using QIIME2 (v. 2020.2) with rarefaction to 13 000 counts, and beta diversity analysis and Boruta feature selection were performed in R using custom R scripts with the packages vegan and Boruta, respectively, after centred log-ratio transformation. Functional profiling of KEGG modules was performed using the SHOGUN functional pipeline based on NCBI's RefSeq release 82.14 Common butyrate-producing genera were identified from existing literature18, 19 and are included in Clostridium clusters IV and XIVa (genera: Clostridium, Ruminococcus, Anaerofilum, Faecalibacterium, Anaerotruncus, Subdoligranulum, Papillibacter, Ruminococcus, Coprococcus, Dorea, Lachnospira, Roseburia, and Butyrivibrio). Raw DNA reads from this study are available at the European Nucleotide Archive under accession number PRJEB63546; three samples each from accessions PRJEB40960 and PRJEB35495 were also included in these analyses.
RESULTS
Patient characteristics
Patient, disease, and transplant characteristics are shown in Table 1. The median (interquartile range) interval between sample collection and cGVHD onset was 233 (184–390) days. We followed the 2014 NIH Consensus Criteria for diagnosis, determining organ involvement and overall severity at diagnosis of cGVHD.1 cGVHD at onset was mild in severity in three patients, moderate in five, and severe in three. Organ distribution and involvement at the onset of cGVHD are shown in Table 1. Of the 11 patients who developed cGVHD, two had overlap syndrome. Type of cGVHD was de-novo in five patients, quiescent in one, and progressive in five. None of the patients had late-onset aGVHD. Of those that developed aGVHD, the maximum grade at onset was 2–4.
| cGVHD | Control | |
|---|---|---|
| Variable, N (%) | N = 11 | N = 13 |
| Gender: Male | 6 (55%) | 11 (85%) |
| Age at transplant | ||
| Median (range) | 59.0 (31–70) | 61.7 (44–70) |
| Donor | ||
| Mismatched-related BM | 1 (9%) | 4 (31%) |
| Matched-related PBSC | 6 (55%) | 4 (31%) |
| UCB | 4 (36%) | 5 (39%) |
| Graft source | ||
| Bone marrow | 1 (9%) | 4 (31%) |
| Periph | 6 (55%) | 4 (31%) |
| Cord | 4 (36%) | 5 (39%) |
| Conditioning | ||
| Myeloablative +TBI | 2 (18%) | 2 (15%) |
| Reduced intensity +TBI | 9 (82%) | 10 (77%) |
| Reduced intensity –TBI | 0 | 1 (8%) |
| GvHD prophylaxis | ||
| CNI/MMF | 10 (91%) | 8 (62%) |
| Other | 1 (9%) | 5 (38%) |
| Disease | ||
| Acute leukaemia | 5 (46%) | 9 (69%) |
| Myelodysplastic syndrome | 2 (18%) | 2 (15%) |
| Multiple myeloma | 0 | 1 (8%) |
| Myeloproliferative disorder | 1 (9%) | 0 |
| NHL | 3 (27%) | 1 (8%) |
| aGVHD | ||
| Grade II–IV | 6 (55%) | 0 |
| Grade III–IV | 4 (36%) | 0 |
| Time to cGVHD | ||
| Median (range), days | 233 (110–705) | - |
| cGVHD NIH global severity at onset | ||
| Mild | 3 (27%) | - |
| Moderate | 5 (46%) | - |
| Severe | 3 (27%) | - |
| cGVHD organ involvement at diagnosis | ||
| Skin | 6 | - |
| Eyes | 6 | - |
| Mouth | 6 | - |
| Liver | 2 | - |
| Gastrointestinal | 3 | - |
| Genitourinary | 0 | - |
| Lung | 1 | - |
| Joints | 3 | - |
| Antibiotic prophylaxis a, b | ||
| Levofloxacin | 10 (91%) | 12 (92%) |
| Penicillin V | 1 (9%) | 0 |
| Cefepime | 0 | 1 (8%) |
| Antibiotics post-HCT (longitudinal sub-cohort) | N = 5 | N = 4 |
| None | 3 (60%) | 1 (25%) |
| Median number of antibiotics prescribed (range) | 4.5 (2–7) | 2 (2–3) |
| Median (range) days post-HCT for antibiotic prescription | 6 (4–8) | 11 (9–12) |
- Abbreviations: aGVHD, acute graft-versus-host disease; BM, bone marrow; cGVHD, chronic graft-versus-host disease; CNI, calcineurin inhibitor; HCV, haematopoietic cell transplant; MMF, mycophenolate mofetil; NHL, non-hodgkin's lymphoma; PBSC, peripheral blood stem cell; TBI, total body irradiation; UCB, umbilical cord blood.
- a Antibiotics prescribed post-HCT included cefepime, metronidazole, vancomycin, levofloxacin, pentamidine, cefpodoxime, and daptomycin.
- b Antibiotics prescribed post-HCT included piperacillin/tazobactam, cefepime, meropenem, vancomycin (iv), and levofloxacin.
Baseline sample analysis
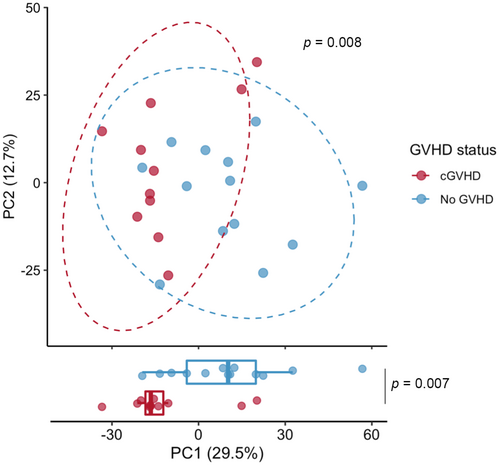
We first examined the overall microbial distribution of the baseline samples in those who subsequently developed cGVHD versus controls. Principal coordinate analysis using the Aitchison distance shows the clustering of baseline cGVHD samples from controls (PERMANOVA p = 0.008, 999 permutations, Figure 1). There was no significant difference in baseline samples' alpha diversity by Shannon Index, Pielou's Evenness, or species richness between the groups.
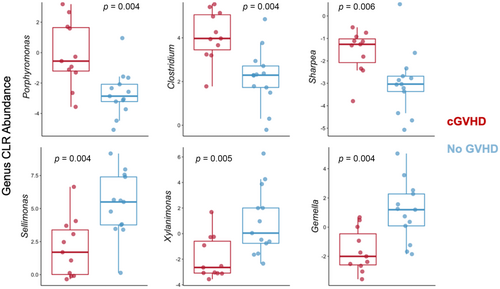
To test whether specific microbial taxa differed between cGVHD and control microbiomes at baseline we used Boruta feature selection, which applies machine learning to weigh the importance of each taxonomic unit in distinguishing the two groups.20 This approach highlighted a significant differential abundance of multiple taxa at baseline between cGVHD and control microbiomes. We noted a lower differential abundance of the phylum Actinobacteria in patients who subsequently developed cGVHD. At the genus level, Clostridium, Porphyromonas and Sharpea had a higher baseline abundance in the cGVHD group; whereas Selimonas, Xylanimonas and Gamella genera had a higher baseline abundance in the control group (Figure 2 of selected taxa, false discovery rate [FDR]-corrected p < 0.01; complete list in Table S1). Multiple species also had significant differential abundance at baseline between the two groups (Figure 3), including a lower differential abundance of Rothia mucilaginous and R. dentocariosa, belonging to the Actinobacteria phylum, in the cGVHD group.
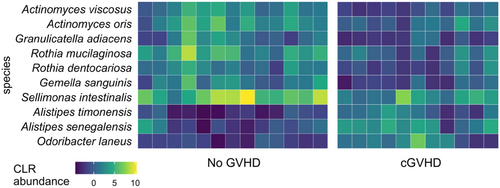
The relative abundance of common butyrate-producing gastrointestinal bacterial genera in baseline samples was similar between the groups with the exception of the Clostridium genus (FDR-corrected p = 0.004), which was present in a higher abundance in baseline samples of those patients who subsequently developed cGVHD.
Functional pathway analysis
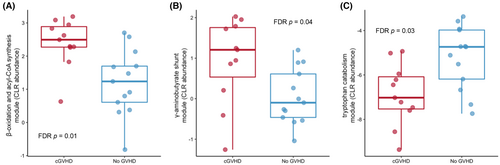
We hypothesized that cGVHD development would follow altered patterns of indole and tryptophan and SCFA pathways, relative to controls. Indeed, Kyoto Encyclopedia of Genes and Genomes (KEGG) functional profiling of baseline sample metagenomes revealed significant differences between controls and cGVHD in genes involved with indole or tryptophan metabolism and in genes affecting SCFA metabolism (Figure 4). Specifically, two pathways associated with SCFA metabolism were enriched in baseline microbiomes of patients who subsequently developed cGVHD: β-oxidation and acyl-CoA synthesis, and the γ-aminobutyrate shunt. In contrast, a tryptophan catabolism pathway was enriched in controls. Those findings could not be confirmed by a simultaneous analysis of plasma or cellular metabolome with no collected samples available for analysis.
Longitudinal sample analysis
Longitudinal changes in the microbiome were compared between baseline and post-engraftment (day 49–130 post-HCT) samples within the same patient, selecting the latest sample available in the pre-specified window when applicable. Paired-sample alpha diversity analysis between groups revealed an overall trend towards a more consistent loss of diversity in patients that subsequently developed cGVHD in both Shannon index and Pielou's Evenness metrics (p = 0.06, paired Wilcoxon test) compared to controls (p ≥ 0.9, paired Wilcoxon test). When looking at the magnitude of within-patient change in alpha diversity, there was a significantly greater loss of alpha diversity (Inverse Simpson, p = 0.016; Pielou's Evenness, p = 0.02; Mann–Whitney U test) in patients from baseline to post-engraftment among those who subsequently developed cGVHD compared to controls (Figures S1A–C ad S2A–C). Post-HCT antibiotic burden was similar between groups (Table 1), though the relatively small sample size prevents a more robust evaluation of this variable.
Given elevated baseline Clostridium genus abundance in cGVHD patients, we compared Clostridium abundance from baseline to post-engraftment between the two groups. Indeed, comparing within-patient Clostridium abundance revealed a significantly greater decline in Clostridium abundance in all patients who subsequently developed cGVHD versus controls (p = 0.016, Mann–Whitney U test; Figures S1D and S2D); interestingly, as opposed to this decrease in Clostridium abundance in cGVHD patients over time, controls consistently showed an increase in Clostridium abundance from baseline to post-engraftment (Figure S1D). When we broadened our search, however, there were no other taxa with significant longitudinal changes over time between groups after multiple-hypothesis correction.
DISCUSSION
Our findings demonstrate previously unreported distinctive patterns in baseline microbiome at the phylum and genus levels and in relevant metabolic pathways, possibly modulating subsequent alloreactivity, in patients who developed cGVHD. Additionally, there was a greater loss of microbial diversity post-engraftment compared to baseline in those subsequently developing cGVHD. The progression to cGVHD may be in part explained by baseline colonization and longitudinal changes in the butyrogenic bacteria Clostridium's abundance as well as functional differences in SCFA and tryptophan metabolism pathways.
Actinobacteria are gram-positive, non-motile, non-spore-forming anaerobic bacteria, and members of this phylum are thought to have key roles in gut barrier homeostasis.21 Lower baseline differential abundance of the phylum Actinobacteria was observed in patients who subsequently develop cGVHD. Therefore, this is consistent with a higher degree of baseline gut barrier damage, predisposing certain patients to subsequent alloreactivity and cGVHD.
At the genus level, Clostridium had a higher baseline differential abundance in the cGVHD group. When we specifically examined the relative abundance of butyrate-producing gastrointestinal bacterial genera, the Clostridium genus was the only one present in a higher abundance in baseline samples in those subsequently developing cGVHD. One possible hypothesis from these observations is that a more severely damaged gut barrier at baseline makes exposure to microbially produced SCFA by elevated Clostridium harmful to colonic stem cells and therefore impairs colonic mucosal recovery.22 While SCFA have been suggested to be beneficial later in recovery,8 our patients who developed cGVHD also had a loss of overall diversity and specifically decreased Clostridium abundance, compared to controls who saw increased Clostridium post engraftment. Together, these data suggest a model for worse recovery outcomes and increasing risk of cGVHD associated with patient microbiome characteristics.7, 22, 23 This further suggests that microbiome-derived signals can regulate and impact immunity differently depending on the specific mucosal environment and epithelial context, and warrants further investigations.24
Our findings should be validated in larger studies with a more homogenous patient population. Additionally, a larger study should investigate the impact of microbiome on developing different types of cGVHD, accounting for previous history of aGVHD. Future studies could additionally investigate the impact of patient, donor, and transplant characteristics on microbiome changes and risk for subsequent GVHD. One limitation of our functional data is we do not have paired plasma samples to correlate functional differences in the microbiome with circulating SCFA levels, bile acid levels, and tryptophan/indole metabolites. Markey et al conducted a case–control study describing a unique post-transplant SCFA and microbiome profile, with no additional insight on the impact of on pre-transplant composition.8 Additionally, correlating our microbiome findings with gut barrier function assays could have further informed our findings. However, we noted an important, significant correlation in functional pathways at baseline, with two pathways associated with SCFA metabolism, namely β-oxidation/acyl-CoA synthesis and γ-aminobutyrate shunt, enriched in the cGVHD patients' microbiomes, while the tryptophan catabolism pathway was enriched in controls. Such metabolites have been shown to play a key role in mediating gut epithelial barrier and mucosal immunity.25-29 A mass spectrometry evaluation of serum samples noted lower leucine and isoleucine with higher cystine in patients subsequently developing cGVHD,9 while another plasma metabolite analysis revealed elevation in plasmas α-ketoglutaric acid before day 100 and at the onset of cGVHD.10 Taken together, those exploratory studies and our findings, highlight the promise of longitudinal pre- and post-transplant microbiome and metabolome analysis to develop future multivariate predictive and preemptive cGVHD biomarker panels.
In summary, we uniquely report here on the association between baseline faecal microbiome composition and metabolic pathways and subsequent cGVHD development in patients undergoing HCT. This work supports the therapeutic potential of microbiome manipulation and metabolic reprograming for cGVHD prevention.
AUTHOR CONTRIBUTIONS
Najla El Jurdi, Shernan G. Holtan and Robin Shields-Cutler contributed to the design; Robin Shields-Cutler performed the analysis; Najla El Jurdi, Shernan G. Holtan and Robin Shields-Cutler analysed the data; Najla El Jurdi and Robin Shields-Cutler performed the bibliographic search and wrote the first version of the manuscript; all other authors edited and revised the manuscript.
ACKNOWLEDGEMENTS
This research received funding from the University of Minnesota.
FUNDING INFORMATION
This research was funded by UMN Grant-in-Aid, Chainbreaker, BRAINS grant (SGH).
CONFLICT OF INTEREST STATEMENT
The authors declare the following conflict of interest, none of which is directly relevant to this work: DJW Research supports FATE, Incyte. BCB, the University of Minnesota, and Moffitt Cancer Center have licensed intellectual property with CRISPR Therapeutics. BCB receives research support from VITRAC Therapeutics. BCB has served on advisory boards for Incyte and CTI BioPharma within the last 36 months. SGH receives research support from Incyte and Vitrac Therapeutics. SGH has spoken at educational programs sponsored by Incyte within the last 36 months.
ETHICS STATEMENT
Study approved by the University of Minnesota Institutional Review Board. Samples were collected from patients who consented to a biorepository protocol.
Open Research
DATA AVAILABILITY STATEMENT
DNA reads are available at the European Nucleotide Archive under accession number PRJEB63546.