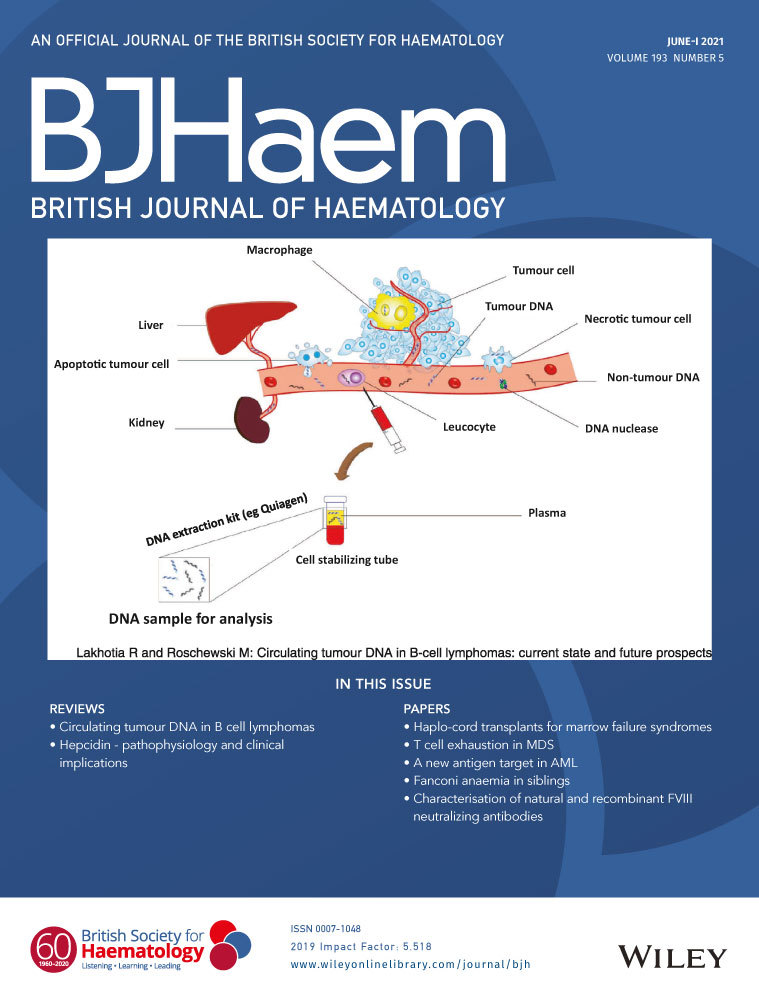
Circulating tumour DNA in B-cell lymphomas: current state and future prospects
Rahul Lakhotia
Lymphoid Malignancies Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD, USA
Search for more papers by this authorCorresponding Author
Mark Roschewski
Lymphoid Malignancies Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD, USA
Correspondence: Mark Roschewski, Lymphoid Malignancies Branch, CCR, NCI, Building 10, Room 4N115, National Institutes of Health, Bethesda, MD 20892, USA.
E-mail: [email protected]
Search for more papers by this authorRahul Lakhotia
Lymphoid Malignancies Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD, USA
Search for more papers by this authorCorresponding Author
Mark Roschewski
Lymphoid Malignancies Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD, USA
Correspondence: Mark Roschewski, Lymphoid Malignancies Branch, CCR, NCI, Building 10, Room 4N115, National Institutes of Health, Bethesda, MD 20892, USA.
E-mail: [email protected]
Search for more papers by this authorSummary
Circulating tumour DNA (ctDNA) is a highly versatile analyte and an emerging biomarker for detection of tumour-specific sequences in lymphoid malignancies. Since ctDNA is derived from tumour cells throughout the body, it overcomes fundamental limitations of tissue biopsies by capturing the complete molecular profile of tumours, including those from inaccessible anatomic locations. Assays for ctDNA are minimally invasive and serial sampling monitors the effectiveness of therapy and identifies minimal residual disease below the detection limit of standard imaging scans. Dynamic changes in ctDNA levels measure real-time tumour kinetics, and early reductions in ctDNA during treatment correlate with clinical outcomes in multiple B-cell lymphomas. After therapy, ctDNA can effectively discriminate between patients who achieved a complete molecular remission from those with residual treatment-resistant disease. Serial monitoring of ctDNA after therapy can detect early molecular relapse and identify drug-resistant clones that harbour targetable mutations. In order for ctDNA to reach its full potential, the standardization and harmonization of the optimal pre-analytical and analytical techniques for B-cell lymphomas is a critically necessary requirement. Prospective validation of ctDNA within clinical studies is also required to determine its clinical utility as an adjunctive decision-making tool.
References
- 1Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001; 61(4): 1659–65.
- 2Stroun M, Anker P, Maurice P, Lyautey J, Lederrey C, Beljanski M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology. 1989; 46(5): 318–22.
- 3Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977; 37(3): 646–50.
- 4Roschewski M, Dunleavy K, Pittaluga S, Moorhead M, Pepin F, Kong K, et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: a correlative biomarker study. Lancet Oncol. 2015; 16(5): 541–9.
- 5Kurtz DM, Scherer F, Jin MC, Soo J, Craig AFM, Esfahani MS, et al. Circulating tumor DNA measurements as early outcome predictors in diffuse large B-cell lymphoma. J Clin Oncol. 2018; 36(28): 2845–53.
- 6Xia L, Li Z, Zhou B, Tian G, Zeng L, Dai H, et al. Statistical analysis of mutant allele frequency level of circulating cell-free DNA and blood cells in healthy individuals. Sci Rep. 2017; 7(1): 7526.
- 7Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. New Engl J Med. 2014; 371(26): 2488–98.
- 8Hu Y, Ulrich BC, Supplee J, Kuang Y, Lizotte PH, Feeney NB, et al. False-positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res. 2018; 24(18): 4437–43.
- 9Rodda AE, Parker BJ, Spencer A, Corrie SR. Extending circulating tumor DNA analysis to ultralow abundance mutations: techniques and challenges. ACS Sens. 2018; 3(3): 540–60.
- 10Kurtz DM, Soo J, Alig S, Keh LCT, Macaulay C, Jin MC, et al. Phased Variant enrichment for enhanced minimal residual disease detection from cell-free DNA. Blood. 2019; 134(Suppl 1): 552.
- 11Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000; 403(6769): 503–11.
- 12Lenz G, Wright GW, Emre NC, Kohlhammer H, Dave SS, Davis RE, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc. Natl Acad. Sci. USA. 2008; 105(36): 13520–5.
- 13Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. New Engl J Med. 2002; 346(25): 1937–47.
- 14Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H, et al. Stromal gene signatures in large-B-cell lymphomas. New Engl J Med. 2008; 359(22): 2313–23.
- 15Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011; 43(9): 830–7.
- 16Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011; 476(7360): 298–303.
- 17Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl Acad. Sci. USA. 2012; 109(10): 3879–84.
- 18Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. New Engl J Med. 2018; 378(15): 1396–407.
- 19Wright GW, Huang DW, Phelan JD, Coulibaly ZA, Roulland S, Young RM, et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell. 2020; 37(4): 551–68 e14.
- 20Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018; 24(5): 679–90.
- 21Araf S, Wang J, Korfi K, Pangault C, Kotsiou E, Rio-Machin A, et al. Genomic profiling reveals spatial intra-tumor heterogeneity in follicular lymphoma. Leukemia. 2018; 32(5): 1261–5.
- 22Hav M, Gerdtsson E, Singh M, Colombo A, Hicks J, Kuhn P, et al. Abstract 2789: highly multiplexed imaging mass cytometry reveals immune cell composition and spatial heterogeneity in diffuse large B cell lymphoma associated with treatment outcome. Cancer Res. 2019; 79: 2789.
- 23Alizadeh AA, Aranda V, Bardelli A, Blanpain C, Bock C, Borowski C, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med. 2015; 21(8): 846–53.
- 24Scherer F, Kurtz DM, Newman AM, Stehr H, Craig AF, Esfahani MS, et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci Transl Med. 2016; 8(364):364ra155.
- 25Rossi D, Diop F, Spaccarotella E, Monti S, Zanni M, Rasi S, et al. Diffuse large B-cell lymphoma genotyping on the liquid biopsy. Blood. 2017; 129(14): 1947–57.
- 26Spina V, Bruscaggin A, Cuccaro A, Martini M, Di Trani M, Forestieri G, et al. Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood. 2018; 131(22): 2413–25.
- 27Suehara Y, Sakata-Yanagimoto M, Hattori K, Nanmoku T, Itoh T, Kaji D, et al. Liquid biopsy for the identification of intravascular large B-cell lymphoma. Haematologica. 2018; 103(6): e241–e244.
- 28Bobillo S, Crespo M, Escudero L, Mayor R, Raheja P, Carpio C, et al. Cell free circulating tumor DNA in cerebrospinal fluid detects and monitors central nervous system involvement of B-cell lymphomas. Haematologica. 2020.
- 29Cheson BD, Fisher RI, Barrington SF, Cavalli F, Schwartz LH, Zucca E, et al. Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: the Lugano classification. J Clin Oncol. 2014; 32(27): 3059–68.
- 30Adams HJ, Nievelstein RA, Kwee TC. Prognostic value of complete remission status at end-of-treatment FDG-PET in R-CHOP-treated diffuse large B-cell lymphoma: systematic review and meta-analysis. Br J Haematol. 2015; 170(2): 185–91.
- 31Melani C, Advani R, Roschewski M, Walters KM, Chen CC, Baratto L, et al. End-of-treatment and serial PET imaging in primary mediastinal B-cell lymphoma following dose-adjusted EPOCH-R: a paradigm shift in clinical decision making. Haematologica. 2018; 103(8): 1337–44.
- 32Schoder H, Polley MY, Knopp MV, Hall NC, Kostakoglu L, Zhang J, et al. Prognostic value of interim FDG-PET in diffuse large cell lymphoma: results from the CALGB 50303 clinical trial. Blood. 2020; 135(25): 2224–2234.
- 33Cheson BD, Ansell S, Schwartz L, Gordon LI, Advani R, Jacene HA, et al. Refinement of the Lugano classification response criteria for lymphoma in the era of immunomodulatory therapy. Blood. 2016; 128(21): 2489–96.
- 34Kurtz DM, Green MR, Bratman SV, Scherer F, Liu CL, Kunder CA, et al. Noninvasive monitoring of diffuse large B-cell lymphoma by immunoglobulin high-throughput sequencing. Blood. 2015; 125(24): 3679–87.
- 35Lakhotia R, Melani C, Pittaluga S, Dunleavy K, Saba NS, Lucas AN, et al. Circulating tumor DNA dynamics during therapy predict outcomes in mantle cell lymphoma. Blood. 2018; 132(Suppl 1): 147.
- 36Greytak SR, Engel KB, Parpart-Li S, Murtaza M, Bronkhorst AJ, Pertile MD, et al. Harmonizing cell-Free DNA collection and processing practices through evidence-based guidance. Clin Cancer Res. 2020; 26(13): 3104–9.
- 37Rossi D, Kurtz DM, Roschewski M, Cavalli F, Zucca E, Wilson WH. The development of liquid biopsy for research and clinical practice in lymphomas: report of the 15-ICML workshop on ctDNA. Hematol Oncol. 2020; 38(1): 34–7.
- 38Lee JS, Kim M, Seong MW, Kim HS, Lee YK, Kang HJ. Plasma vs. serum in circulating tumor DNA measurement: characterization by DNA fragment sizing and digital droplet polymerase chain reaction. Clin Chem Lab Med. 2020; 58(4): 527–32.
- 39Armand P, Oki Y, Neuberg DS, Faham M, Cummings C, Klinger M, et al. Detection of circulating tumour DNA in patients with aggressive B-cell non-Hodgkin lymphoma. Br J Haematol. 2013; 163(1): 123–6.
- 40Oki Y, Neelapu SS, Fanale M, Kwak LW, Fayad L, Rodriguez MA, et al. Detection of classical Hodgkin lymphoma specific sequence in peripheral blood using a next-generation sequencing approach. Br J Haematol. 2015; 169(5): 689–693.
- 41Kang Q, Henry NL, Paoletti C, Jiang H, Vats P, Chinnaiyan AM, et al. Comparative analysis of circulating tumor DNA stability In K3EDTA, Streck, and Cell Save blood collection tubes. Clin Biochem. 2016; 49(18): 1354–60.
- 42Dreyer WJ, Bennett JC. The molecular basis of antibody formation: a paradox. Proc Natl Acad Sci U S A. 1965; 54(3): 864–9.
- 43Tonegawa S. Somatic generation of antibody diversity. Nature. 1983; 302(5909): 575–81.
- 44Pott C, Hoster E, Delfau-Larue MH, Beldjord K, Bottcher S, Asnafi V, et al. Molecular remission is an independent predictor of clinical outcome in patients with mantle cell lymphoma after combined immunochemotherapy: a European MCL intergroup study. Blood. 2010; 115(16): 3215–23.
- 45Evans PA, Pott C, Groenen PJ, Salles G, Davi F, Berger F, et al. Significantly improved PCR-based clonality testing in B-cell malignancies by use of multiple immunoglobulin gene targets. Report of the BIOMED-2 Concerted Action BHM4-CT98-3936. Leukemia. 2007; 21(2): 207–14.
- 46Gibson UE, Heid CA, Williams PM. A novel method for real time quantitative RT-PCR. Genome Res. 1996; 6(10): 995–1001.
- 47Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996; 6(10): 986–94.
- 48Pott C, Bruggemann M, Ritgen M, van der Velden VHJ, van Dongen JJM, Kneba M. MRD detection in B-cell non-hodgkin lymphomas using Ig gene rearrangements and chromosomal translocations as targets for real-time quantitative PCR. Methods Mol Biol. 2019; 1956: 199–228.
- 49Limpens J, Stad R, Vos C, de Vlaam C, de Jong D, van Ommen GJ, et al. Lymphoma-associated translocation t(14;18) in blood B cells of normal individuals. Blood. 1995; 85(9): 2528–36.
- 50Dolken G, Illerhaus G, Hirt C, Mertelsmann R. BCL-2/JH rearrangements in circulating B cells of healthy blood donors and patients with nonmalignant diseases. J Clin Oncol. 1996; 14(4): 1333–44.
- 51Hudecova I. Digital PCR analysis of circulating nucleic acids. Clin Biochem. 2015; 48(15): 948–56.
- 52Camus V, Sarafan-Vasseur N, Bohers E, Dubois S, Mareschal S, Bertrand P, et al. Digital PCR for quantification of recurrent and potentially actionable somatic mutations in circulating free DNA from patients with diffuse large B-cell lymphoma. Leuk Lymphoma. 2016; 57(9): 2171–9.
- 53Drandi D, Alcantara M, Benmaad I, Sohlbrandt A, Lhermitte L, Zaccaria G, et al. Droplet digital PCR quantification of mantle cell lymphoma follow-up samples from four prospective trials of the European MCL network. Hemasphere. 2020; 4(2): e347.
- 54Hiemcke-Jiwa LS, Minnema MC, Radersma-van Loon JH, Jiwa NM, de Boer M, Leguit RJ, et al. The use of droplet digital PCR in liquid biopsies: a highly sensitive technique for MYD88 p.(L265P) detection in cerebrospinal fluid. Hematol Oncol. 2018; 36(2): 429–435.
- 55Ladetto M, Bruggemann M, Monitillo L, Ferrero S, Pepin F, Drandi D, et al. Next-generation sequencing and real-time quantitative PCR for minimal residual disease detection in B-cell disorders. Leukemia. 2014; 28(6): 1299–307.
- 56Faham M, Zheng J, Moorhead M, Carlton VE, Stow P, Coustan-Smith E, et al. Deep-sequencing approach for minimal residual disease detection in acute lymphoblastic leukemia. Blood. 2012; 120(26): 5173–80.
- 57Herrera AF, Armand P. Minimal residual disease assessment in lymphoma: methods and applications. J Clin Oncol. 2017; 35(34): 3877–87.
- 58Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011; 108(23): 9530–5.
- 59Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014; 20(5): 548–54.
- 60Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016; 34(5): 547–55.
- 61Bohers E, Viailly PJ, Dubois S, Bertrand P, Maingonnat C, Mareschal S, et al. Somatic mutations of cell-free circulating DNA detected by next-generation sequencing reflect the genetic changes in both germinal center B-cell-like and activated B-cell-like diffuse large B-cell lymphomas at the time of diagnosis. Haematologica. 2015; 100(7): e280–e284.
- 62Liu J, Chen X, Wang J, Zhou S, Wang CL, Ye MZ, et al. Biological background of the genomic variations of cf-DNA in healthy individuals. Ann Oncol. 2019; 30(3): 464–70.
- 63Abbosh C, Swanton C, Birkbak NJ. Clonal haematopoiesis: a source of biological noise in cell-free DNA analyses. Ann Oncol. 2019; 30(3): 358–9.
- 64Chiu BC, Zhang Z, You Q, Zeng C, Stepniak E, Bracci PM, et al. Prognostic implications of 5-hydroxymethylcytosines from circulating cell-free DNA in diffuse large B-cell lymphoma. Blood Adv. 2019; 3(19): 2790–9.
- 65Chambwe N, Kormaksson M, Geng H, De S, Michor F, Johnson NA, et al. Variability in DNA methylation defines novel epigenetic subgroups of DLBCL associated with different clinical outcomes. Blood. 2014; 123(11): 1699–708.
- 66Song CX, Yin S, Ma L, Wheeler A, Chen Y, Zhang Y, et al. 5-Hydroxymethylcytosine signatures in cell-free DNA provide information about tumor types and stages. Cell Res. 2017; 27(10): 1231–42.
- 67Moss J, Magenheim J, Neiman D, Zemmour H, Loyfer N, Korach A, et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun. 2018; 9(1): 5068.
- 68Shen SY, Singhania R, Fehringer G, Chakravarthy A, Roehrl MHA, Chadwick D, et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature. 2018; 563(7732): 579–83.
- 69Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell. 2016; 164(1–2): 57–68.
- 70Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018; 173(2): 291–304 e6.
- 71Raman LVDLM, Vriendt CD, Van Den Broeck B, Muylle K, Deeren D, Menten B, et al. Exploring the potential of shallow whole-genome sequencing for diagnosis and disease monitoring of lymphoma in liquid biopsy. Blood. 2019; 134(Suppl 1): 2087.
- 72Johl A, Lengfelder E, Hiddemann W, Klapper W; German Low-grade Lymphoma Study Group. Core needle biopsies and surgical excision biopsies in the diagnosis of lymphoma-experience at the Lymph Node Registry Kiel. Ann Hematol. 2016; 95(8): 1281–6.
- 73Khatab S, Spliet W, Woerdeman PA. Frameless image-guided stereotactic brain biopsies: emphasis on diagnostic yield. Acta Neurochir (Wien). 2014; 156(8): 1441–50.
- 74Onder E, Arikok AT, Onder S, Han U, Sorar M, Kertmen H, et al. Corticosteroid pre-treated primary CNS lymphoma: a detailed analysis of stereotactic biopsy findings and consideration of interobserver variability. Int J Clin Exp Pathol. 2015; 8(7): 7798–808.
- 75Pittman M, Treese S, Chen L, Frater JL, Nguyen TT, Hassan A, et al. Utility of flow cytometry of cerebrospinal fluid as a screening tool in the diagnosis of central nervous system lymphoma. Arch Pathol Lab Med. 2013; 137(11): 1610–8.
- 76Hickmann AK, Frick M, Hadaschik D, Battke F, Bittl M, Ganslandt O, et al. Molecular tumor analysis and liquid biopsy: a feasibility investigation analyzing circulating tumor DNA in patients with central nervous system lymphomas. BMC Cancer. 2019; 19(1): 192.
- 77Grommes C, Tang SS, Wolfe J, Kaley TJ, Daras M, Pentsova EI, et al. Phase 1b trial of an ibrutinib-based combination therapy in recurrent/refractory CNS lymphoma. Blood. 2019; 133(5): 436–45.
- 78Chapuy B, Roemer MG, Stewart C, Tan Y, Abo RP, Zhang L, et al. Targetable genetic features of primary testicular and primary central nervous system lymphomas. Blood. 2016; 127(7): 869–81.
- 79Hiemcke-Jiwa LS, Leguit RJ, Snijders TJ, Bromberg JEC, Nierkens S, Jiwa NM, et al. MYD88 p. (L265P) detection on cell-free DNA in liquid biopsies of patients with primary central nervous system lymphoma. Br J Haematol. 2019; 185(5): 974–7.
- 80Fontanilles M, Marguet F, Bohers E, Viailly PJ, Dubois S, Bertrand P, et al. Non-invasive detection of somatic mutations using next-generation sequencing in primary central nervous system lymphoma. Oncotarget. 2017; 8(29): 48157–68.
- 81Ponzoni M, Ferreri AJ, Campo E, Facchetti F, Mazzucchelli L, Yoshino T, et al. Definition, diagnosis, and management of intravascular large B-cell lymphoma: proposals and perspectives from an international consensus meeting. J Clin Oncol. 2007; 25(21): 3168–73.
- 82Ruppert AS, Dixon JG, Salles GA, Wall A, Cunningham D, Poeschel V, et al. International prognostic indices in diffuse large B-cell lymphoma (DLBCL): a comparison of IPI, R-IPI and NCCN-IPI. Blood. 2020' 135(23): 2041–8.
- 83Kanoun S, Rossi C, Berriolo-Riedinger A, Dygai-Cochet I, Cochet A, Humbert O, et al. Baseline metabolic tumour volume is an independent prognostic factor in Hodgkin lymphoma. Eur J Nucl Med Mol Imaging. 2014; 41(9): 1735–43.
- 84Sasanelli M, Meignan M, Haioun C, Berriolo-Riedinger A, Casasnovas RO, Biggi A, et al. Pretherapy metabolic tumour volume is an independent predictor of outcome in patients with diffuse large B-cell lymphoma. Eur J Nucl Med Mol Imaging. 2014; 41(11): 2017–22.
- 85Meignan M, Cottereau AS, Versari A, Chartier L, Dupuis J, Boussetta S, et al. Baseline metabolic tumor volume predicts outcome in high-tumor-burden follicular lymphoma: a pooled analysis of three multicenter studies. J Clin Oncol. 2016; 34(30): 3618–26.
- 86Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016; 127(20): 2375–90.
- 87Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004; 103(1): 275–82.
- 88Kurtz DM, Esfahani MS, Scherer F, Soo J, Jin MC, Liu CL, et al. Dynamic risk profiling using serial tumor biomarkers for personalized outcome prediction. Cell. 2019; 178(3): 699–713 e19.
- 89Lakhotia R, Melani C, Pittaluga S, Dunleavy K, Saba NS, Lucas AN, et al. Circulating tumor DNA dynamics during therapy predict outcomes in mantle cell lymphoma. Blood. 2018; 132(Suppl 1): 147.
- 90Smith M, Jegede O, Parekh S, Hanson CA, Martin P, Till BG, et al. Minimal Residual Disease (MRD) Assessment in the ECOG1411 Randomized Phase 2 Trial of Front-Line Bendamustine-Rituximab (BR)-Based Induction Followed By Rituximab (R)±Lenalidomide (L) Consolidation for Mantle Cell Lymphoma (MCL). Blood. 2019; 134(Suppl 1): 751.
- 91Mathas S, Hartmann S, Kuppers R. Hodgkin lymphoma: pathology and biology. Semin Hematol. 2016; 53(3): 139–47.
- 92Camus V, Stamatoullas A, Mareschal S, Viailly PJ, Sarafan-Vasseur N, Bohers E, et al. Detection and prognostic value of recurrent exportin 1 mutations in tumor and cell-free circulating DNA of patients with classical Hodgkin lymphoma. Haematologica. 2016; 101(9): 1094–101.
- 93Camus V, Viennot M, Lequesne J, Viailly PJ, Bohers E, Bessi L, et al. Targeted genotyping of circulating tumor DNA for classical Hodgkin Lymphoma monitoring: a prospective study. Haematologica. 2020.
- 94Sarkozy C, Huet S, Carlton VE, Fabiani B, Delmer A, Jardin F, et al. The prognostic value of clonal heterogeneity and quantitative assessment of plasma circulating clonal IG-VDJ sequences at diagnosis in patients with follicular lymphoma. Oncotarget. 2017; 8(5): 8765–74.
- 95Delfau-Larue MH, van der Gucht A, Dupuis J, Jais JP, Nel I, Beldi-Ferchiou A, et al. Total metabolic tumor volume, circulating tumor cells, cell-free DNA: distinct prognostic value in follicular lymphoma. Blood Adv. 2018; 2(7): 807–16.
- 96Pott C, Knecht H, Herzog A, Genuardi E, Unterhalt M, Mantoan B, et al. Standardized IGH-based next-generation sequencing for MRD detection in follicular lymphoma. Blood. 2017; 130(Suppl 1): 1491.
- 97Delfau-Larue MH, Boulland ML, Beldi-Ferchiou A, Feugier P, Maisonneuve H, Casasnovas RO, et al. Lenalidomide/rituximab induces high molecular response in untreated follicular lymphoma: LYSA ancillary RELEVANCE study. Blood Adv. 2020; 4(14): 3217–23.
- 98Di Giacomo AM, Danielli R, Guidoboni M, Calabro L, Carlucci D, Miracco C, et al. Therapeutic efficacy of ipilimumab, an anti-CTLA-4 monoclonal antibody, in patients with metastatic melanoma unresponsive to prior systemic treatments: clinical and immunological evidence from three patient cases. Cancer Immunol Immunother. 2009; 58(8): 1297–306.
- 99Hossain NM, Dahiya S, Le R, Abramian AM, Kong KA, Muffly LS, et al. Circulating tumor DNA assessment in patients with diffuse large B-cell lymphoma following CAR T-cell therapy. Leuk Lymphoma. 2019; 60(2): 503–6.
- 100Frank MJ, Hossain N, Bukhari A, Dean E, Spiegel JY, Claire GK, et al. Detectable circulating tumor DNA 28 days after the CD19 CAR T-cell therapy, axicabtagene ciloleucel, is associated with poor outcomes in patients with diffuse large B-cell lymphoma. Blood. 2019; 134(Suppl 1): 884.
- 101Goldschmidt N, Or O, Klein M, Savitsky B, Paltiel O. The role of routine imaging procedures in the detection of relapse of patients with Hodgkin lymphoma and aggressive non-Hodgkin lymphoma. Ann Hematol. 2011; 90(2): 165–71.
- 102Thompson CA, Ghesquieres H, Maurer MJ, Cerhan JR, Biron P, Ansell SM, et al. Utility of routine post-therapy surveillance imaging in diffuse large B-cell lymphoma. J Clin Oncol. 2014; 32(31): 3506–12.
- 103Huntington SF, Svoboda J, Doshi JA. Cost-effectiveness analysis of routine surveillance imaging of patients with diffuse large B-cell lymphoma in first remission. J Clin Oncol. 2015; 33(13): 1467–74.
- 104Hamlin PA, Zelenetz AD, Kewalramani T, Qin J, Satagopan JM, Verbel D, et al. Age-adjusted International Prognostic Index predicts autologous stem cell transplantation outcome for patients with relapsed or primary refractory diffuse large B-cell lymphoma. Blood. 2003; 102(6): 1989–96.
- 105Park JH, Riviere I, Gonen M, Wang X, Senechal B, Curran KJ, et al. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N Engl J Med. 2018; 378(5): 449–59.
- 106Cohen JB, Kurtz DM, Staton AD, Flowers CR. Next-generation surveillance strategies for patients with lymphoma. Future Oncol. 2015; 11(13): 1977–91.
- 107Rushton CK, Arthur SE, Alcaide M, Cheung M, Jiang A, Coyle KM, et al. Genetic and evolutionary patterns of treatment resistance in relapsed B-cell lymphoma. Blood Adv. 2020; 4(13): 2886–98.
- 108Al-Tourah AJ, Gill KK, Chhanabhai M, Hoskins PJ, Klasa RJ, Savage KJ, et al. Population-based analysis of incidence and outcome of transformed non-Hodgkin's lymphoma. J Clin Oncol. 2008; 26(32): 5165–9.
- 109Casulo C, Friedberg J. Transformation of marginal zone lymphoma (and association with other lymphomas). Best Pract Res Clin Haematol. 2017; 30(1–2): 131–8.
- 110Durot E, Tomowiak C, Michallet AS, Dupuis J, Lepretre S, Toussaint E, et al. Retrospective analysis of 56 cases of transformed Waldenström macroglobulinemia. A study on behalf of the French Innovative Leukemia Organization (FILO). Blood. 2016; 128(22): 2982.
- 111Pasqualucci L, Khiabanian H, Fangazio M, Vasishtha M, Messina M, Holmes AB, et al. Genetics of follicular lymphoma transformation. Cell Rep. 2014; 6(1): 130–40.
- 112Kridel R, Chan FC, Mottok A, Boyle M, Farinha P, Tan K, et al. Histological transformation and progression in follicular lymphoma: a clonal evolution study. PLoS Med. 2016; 13(12):e1002197.
- 113Wilson WH, Young RM, Schmitz R, Yang Y, Pittaluga S, Wright G, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med. 2015; 21(8): 922–6.
- 114Martin P, Maddocks K, Leonard JP, Ruan J, Goy A, Wagner-Johnston N, et al. Postibrutinib outcomes in patients with mantle cell lymphoma. Blood. 2016; 127(12): 1559–63.
- 115Rahal R, Frick M, Romero R, Korn JM, Kridel R, Chan FC, et al. Pharmacological and genomic profiling identifies NF-kappaB-targeted treatment strategies for mantle cell lymphoma. Nat Med. 2014; 20(1): 87–92.
- 116Agarwal R, Chan YC, Tam CS, Hunter T, Vassiliadis D, Teh CE, et al. Dynamic molecular monitoring reveals that SWI-SNF mutations mediate resistance to ibrutinib plus venetoclax in mantle cell lymphoma. Nat Med. 2019; 25(1): 119–29.
- 117Huw LY, O'Brien C, Pandita A, Mohan S, Spoerke JM, Lu S, et al. Acquired PIK3CA amplification causes resistance to selective phosphoinositide 3-kinase inhibitors in breast cancer. Oncogenesis. 2013; 2: e83.
- 118Nakanishi Y, Walter K, Spoerke JM, O'Brien C, Huw LY, Hampton GM, et al. Activating mutations in PIK3CB confer resistance to PI3K inhibition and define a novel oncogenic role for p110beta. Cancer Res. 2016; 76(5): 1193–203.
- 119Castellano E, Downward J. RAS interaction with PI3K: more than just another effector pathway. Genes Cancer. 2011; 2(3): 261–74.
- 120Hoy SM. Tazemetostat: first approval. Drugs. 2020; 80(5): 513–21.
- 121Morschhauser F, Tilly H, Chaidos A, Phillips TJ, Ribrag V, Campbell P, et al. Phase 2 multicenter study of tazemetostat, an EZH2 inhibitor, in patients with relapsed or refractory follicular lymphoma. Blood. 2019; 134(Suppl 1): 123.
- 122Daigle S, McDonald AA, Morschhauser F, Salles G, Ribrag V, McKay P, et al. Discovery of candidate predictors of response to tazemetostat in diffuse large B-cell lymphoma and follicular lymphoma using NGS technology on ctDNA samples collected pre-treatment. Blood. 2017; 130(Suppl 1): 4013.
- 123Hiemcke-Jiwa LS, Ten Dam-van Loon NH, Leguit RJ, Nierkens S, Ossewaarde-van Norel J, de Boer JH, et al. Potential diagnosis of vitreoretinal lymphoma by detection of MYD88 mutation in aqueous humor with ultrasensitive droplet digital polymerase chain reaction. JAMA Ophthalmol. 2018; 136(10): 1098–104.
- 124Cani AK, Hovelson DH, Demirci H, Johnson MW, Tomlins SA, Rao RC. Next generation sequencing of vitreoretinal lymphomas from small-volume intraocular liquid biopsies: new routes to targeted therapies. Oncotarget. 2017; 8(5): 7989–98.
- 125Bohers E, Viailly PJ, Becker S, Marchand V, Ruminy P, Maingonnat C, et al. Non-invasive monitoring of diffuse large B-cell lymphoma by cell-free DNA high-throughput targeted sequencing: analysis of a prospective cohort. Blood Cancer J. 2018; 8(8): 74.
- 126Assouline SE, Nielsen TH, Yu S, Alcaide M, Chong L, MacDonald D, et al. Phase 2 study of panobinostat with or without rituximab in relapsed diffuse large B-cell lymphoma. Blood. 2016; 128(2): 185–94.
- 127Roschewski MJ, Melani CJ, Pittaluga S, Dunleavy K, Saba NS, Grant C, et al. Circulating tumor DNA to predict timing of relapse in mantle cell lymphoma. J Clin Oncol. 2018; 36(Suppl 15): 7576.