The association of four genetic variants with myelosuppression in gemcitabine-treated Japanese is not evident in gemcitabine/carboplatin-treated Swedes
Funding information: Marcus Borgströms stiftelse; Funds of Radiumhemmet; ALF grants Region Östergötland; Linköping University; Swedish Research Council; Swedish Cancer Society
Abstract
Gemcitabine/carboplatin-induced myelosuppressive adverse drug reactions (ADRs) are clinical problems leading to patient suffering and dose alterations. There is a need for personalised medicine to improve treatment effects and patients' well-being. We tested four genetic variants, rs11141915, rs1901440, rs12046844 and rs11719165, previously suggested as potential biomarkers for gemcitabine-induced leukopenia/neutropenia in Japanese patients, in 213 Swedish gemcitabine/carboplatin-treated non-small cell lung cancer (NSCLC) patients. DNA was genotyped using TaqMan probes and real-time PCR. The relationships between the risk alleles and low toxicity (non-ADR: Common Terminology Criteria for Adverse Events [CTCAE] grades 0) or high toxicity (ADR: CTCAE grades 3–4) of platelets, leukocytes and neutrophils were evaluated using Fisher's exact test. The risk alleles did not correlate with myelosuppression, and the strongest borderline significance (not withstanding adjustment for multiple testing) was for rs1901440 (neutropenia, p = 0.043) and rs11719165 (leukopenia, p = 0.049) where the risk alleles trended towards lower toxicity, contrasting with previous study findings. Risk alleles and higher risk scores were more common among our patients. We conclude that the genetic variants do not apply to Swedish patients treated with gemcitabine/carboplatin. However, they can still be important in other populations and cohorts, especially in a gemcitabine monotherapy setting, where the causal genetic variation might influence myelosuppressive ADRs.
1 INTRODUCTION
It is well known that cancer treatments are harsh and that the worse the prognosis, the harsher these treatments tend to be. The set of available treatment options is rapidly improving and growing, now including surgery, radiation, classical chemotherapy, immune checkpoint inhibitors and targeted drugs. The classical chemotherapies are needed and used as they still have an important role in the treatment of cancer, although decreasingly as the first-line choice. Gemcitabine and carboplatin are two classical chemotherapies that are used in various combinations to combat cancer. Both their individual and combined uses are associated with the induction of severe adverse drug reactions (ADRs),1-3 which can be dose-limiting and result in reduced, delayed or interrupted treatments. Specifically, myelosuppression in the form of thrombocytopenia, leukopenia and neutropenia is commonly induced by both gemcitabine and carboplatin, and around 50% of treated patients experience Common Terminology Criteria for Adverse Events (CTCAE) grades 3–4 (i.e., severe to life-threatening ADRs).1-11 There is a need for more personalised treatment regimens to improve both treatment outcomes and patients' well-being.12, 13
Kiyotani et al.14 performed a genome-wide association study (GWAS) and identified four genetic variants, rs11141915, rs1901440, rs12046844 and rs11719165, that were associated with gemcitabine-induced leukopenia/neutropenia, especially in patients having two to three risk genotypes. Low et al.15 then found further associations between gemcitabine-induced myelosuppressive toxicity for three of the four genetic variants reported by Kiyotani et al.14: rs1901440 (via linkage disequilibrium [LD] with rs6430443), rs12046844 and rs11719165. However, as pointed out by the authors in Low et al.,15 these two studies are, to some extent, based on the same patient material.
In this study, we aimed to test whether the four proposed genetic variants associated with myelosuppressive ADRs could also be used for risk assessment in our cohort of Swedish gemcitabine/carboplatin-treated non-small cell lung cancer (NSCLC) patients.
2 MATERIALS AND METHODS
2.1 Patients
A total of 215 NSCLC patients had been recruited at the Karolinska University Hospital in Stockholm, Sweden, after giving informed consent as per the Helsinki Declaration. The study was approved by the regional ethics committee in Stockholm, Sweden (DNR-03-413 with amendment 2016/258-32/1). The study was conducted in accordance with the Basic & Clinical Pharmacology & Toxicology policy for experimental and clinical studies.16 This material has been used in other toxicity studies.8-11 All patients received at least one cycle of carboplatin (AUC = 5, on Day 1) and gemcitabine (1250 mgm−2, on Day 1 and Day 8). For this study, 213 of the included patients were included.
2.2 DNA extraction and genotyping
DNA was extracted from blood samples using the QIAamp DNA Mini Kit (Qiagen, Venlo, Netherlands) according to the manufacturer's protocol. Genotyping of rs11141915, rs1901440, rs12046844 and rs11719165 was carried out according to the manufacturer's protocols using the TaqMan SNP Genotyping Assay (Applied Biosystems, Carlsbad, CA, USA) listed in Table 1, the TaqMan Genotyping Master Mix (Applied Biosystems) and the 7900HT Fast Real-Time PCR System (Applied Biosystems) using standard mode thermal cycling conditions. To confirm the robustness of the genotyping, 20 of the samples were randomly selected and genotyped twice using the same method and one sample determined heterozygous for each genotype was sent to GATC Biotech AG, European Custom Sequencing Centre, Cologne, Germany, for Sanger sequencing for genotype verification.
| rsID | Chr | Base paira | Gene | Consequence | Reporter alleles [VIC/FAM] in assay | AssayID |
|---|---|---|---|---|---|---|
| rs11141915 | 9 | 90,235,794 | DAPK1 | Intron | T/Gb | C___1387009_10 |
| rs1901440 | 2 | 134,437,959 | - | Intergenic | A/C | C___1356323_10 |
| rs12046844 | 1 | 66,238,379 | - | Intergenic | C/Tc | AHLJ2MD |
| rs11719165 | 3 | 194,586,088 | - | Intergenic | C/T | C__31058029_20 |
- Note: VIC and FAM are the two fluorescent dyes used to discriminate between the alleles. AssayIDs used by Applied Biosystems.
- a Base pair in assembly GRCh37.
- b Converted from A/C as reported by VIC/FAM to facilitate comparison with the previous studies.
- c Converted from G/A as reported by VIC/FAM to facilitate comparison with the previous studies.
2.3 Myelosuppression
Platelet, leukocyte and neutrophil counts were registered at baseline and Days 8, 15 and 21 of the first cycle. The nadir values during the first cycle were graded according to National Cancer Institute (NCI) CTCAE Version 4.03 and used as the toxicity endpoints for platelets, leukocytes and neutrophils, respectively. Also, the maximum toxicity of leukocytes and neutrophils (Max Leu/Neu) was evaluated as a toxicity endpoint.
2.4 HapMap and LD data
Genotype frequencies for the four genetic variants from HapMap-CEU and HapMap-JPT datasets on dbSNP17 (https://www.ncbi.nlm.nih.gov/snp/) were retrieved on 2020-11-15, listed under the ss#: rs11141915, ss43792244; rs1901440, ss78174812; rs12046844, ss87503038; and rs11719165, ss44412686. LD plots of the regions around the genetic variants were generated using Haploview Version 4.218 and 1000 genomes phase 3 LD data in the region ±10,000 base pairs of the respective genetic variants for both CEU and JPT populations from Ensembl GRCh37.p13 release 10219 (http://grch37.ensembl.org/Homo_sapiens/Tools/VcftoPed).
2.5 Statistics
All statistics were calculated using the statistical environment R Version 4.0.3.20 The 213 patients were split into two groups, ADR (CTCAE grades 3–4) and non-ADR (CTCAE grades 0), based on all toxicity endpoints as in the original study by Kiyotani et al.,14 meaning that patients with intermediate toxicity (CTCAE grades 1–2) were excluded. The allele frequencies of the four genetic variants were then compared between the ADR and non-ADR groups using allelic, dominant and recessive Fisher's exact tests21 based on the proposed risk alleles. Each patient was then scored according to the number of sites that were homozygous for the risk alleles, that is, using the same risk scoring system as Kiyotani et al.14 This gives a score between 0 and 4 for each patient. The proportion of patients with various risk scores in the ADR and non-ADR groups were compared using Fisher's exact test, to evaluate further if the combination of the genetic variants could be used for predicting the risk of gemcitabine/carboplatin-induced myelosuppression. Bonferroni correction was implemented to test if significant p-values surpass adjustments for multiple testing.
3 RESULTS
3.1 Patient characteristics
The clinical characteristics of the 213 NSCLC patients in this study are presented in Table 2, and the overall myelosuppressive toxicities, graded according to the CTCAE, are shown in Table 3. From this, it is evident that high grades (3–4) are common and experienced by 35%, 23%, 45% and 48% of patients, respectively, for platelets, leukocytes, neutrophils and Max Leu/Neu, during the first treatment cycle.
| Age, in years, median (range) | 65 | (45–83) |
| Gender, N (%) | ||
| Female | 113 | 53.1% |
| Male | 100 | 46.9% |
| Smoking, N (%) | ||
| Current | 94 | 44.1% |
| Former | 98 | 46.0% |
| Never | 21 | 9.9% |
| Histological type, N (%) | ||
| Adenocarcinoma (AC) | 131 | 61.5% |
| Squamous cell carcinomas (SCC) | 40 | 18.8% |
| Non-small cell lung cancer (NSCLC) | 31 | 14.6% |
| Large cell carcinoma (LLC) | 10 | 4.7% |
| Not specified | 1 | 0.5% |
| Pathological stage, N (%) | ||
| I | 40 | 18.8% |
| II | 28 | 13.1% |
| III | 63 | 29.6% |
| IV | 80 | 37.6% |
| Not specified | 2 | 0.9% |
| CTCAE grades | 0 | 1 | 2 | 3 | 4 | Not specified | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Platelets | 44 | 20.7% | 50 | 23.5% | 44 | 20.7% | 44 | 20.7% | 30 | 14.1% | 1 | 0.5% |
| Leukocytes | 64 | 30.0% | 28 | 13.1% | 72 | 33.8% | 44 | 20.7% | 5 | 2.3% | 0 | 0.0% |
| Neutrophils | 73 | 34.3% | 4 | 1.9% | 23 | 10.8% | 63 | 29.6% | 33 | 15.5% | 17 | 8.0% |
| Max Leu/Neu | 59 | 27.7% | 14 | 6.6% | 37 | 17.4% | 68 | 31.9% | 35 | 16.4% | 0 | 0.0% |
- Note: Also shown is the maximum toxicity of leukocytes and neutrophils (Max Leu/Neu).
3.2 Genotyping, HapMap and LD
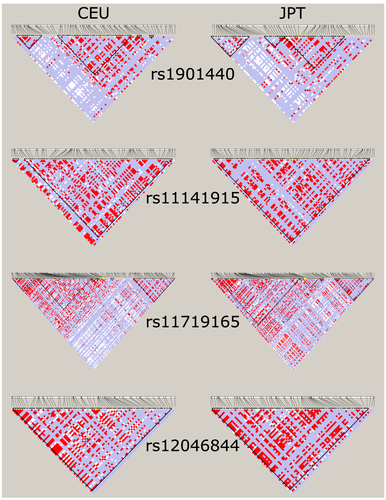
The genotyping of the four genetic variants using TaqMan probes was successful in all but one sample for which rs12046844 and rs11719165 were undeterminable. All genotype frequencies are listed in Table 4. The 20 randomly selected samples showed the same results in the replication round, and the selected heterozygous samples were also confirmed as heterozygous using Sanger sequencing (GATC Biotech AG, European Custom Sequencing Centre, Cologne, Germany). Furthermore, the genotypes in this cohort are not out of the Hardy Weinberg equilibrium. The presented genotype frequencies follow distributions similar to the HapMap-CEU cohort (Table 4). However, compared with HapMap-JPT (Table 4), there are some differences in the genotype distributions, especially for rs12046844 and rs11719165 where the risk alleles (C in both cases) are more common in the patient cohort presented here and the HapMap-CEU data compared with the HapMap-JPT data. From the LD plots of the regions around the genetic variants (±10,000 base pairs) generated using Haploview and 1000 genomes phase 3 data for CEU and JPT populations, shown in Figure 1, it is evident that the LD structure differs in the underlying populations used in the present study compared with the two previous studies by Kiyotani et al.14 and Low et al.15
| Genotype | N | % | HapMap-CEU | HapMap-JPT |
|---|---|---|---|---|
| rs11141915 | ||||
| T/T | 125 | 58.7% | 58.4% | 43.0% |
| T/G | 74 | 34.7% | 38.1% | 45.3% |
| G/G | 14 | 6.6% | 3.5% | 11.6% |
| Undetermined | 0 | - | - | - |
| rs1901440 | ||||
| A/A | 98 | 46.0% | 45.9% | 60.0% |
| A/C | 94 | 44.1% | 45.0% | 37.6% |
| C/C | 21 | 9.9% | 9.2% | 2.4% |
| Undetermined | 0 | - | - | - |
| rs12046844 | ||||
| C/C | 130 | 61.0% | 69.9% | 34.9% |
| C/T | 75 | 35.2% | 28.3% | 48.8% |
| T/T | 7 | 3.3% | 1.8% | 16.3% |
| Undetermined | 1 | 0.5% | - | - |
| rs11719165 | ||||
| C/C | 86 | 40.4% | 30.5% | 17.8% |
| C/T | 104 | 48.8% | 50.8% | 44.4% |
| T/T | 22 | 10.3% | 18.6% | 37.8% |
| Undetermined | 1 | 0.5% | - | - |
- Note: The genotype of one sample was undeterminable for both rs12046844 and rs11719165.
3.3 Genetic variants and toxicity
The results from all association tests between genotypes and myelosuppressive toxicities are listed in Table 5. Overall, the associations were mainly non-significant, and for many of the toxicity endpoints, the risk allele frequencies were similar in the ADR and non-ADR groups and for some even higher in the non-ADR groups. Two of the tests yielded borderline significant results, rs1901440 for neutrophils (p = 0.043, odds ratio [OR] = 0.52, 95% confidence interval [CI] = 0.26–1.01) and rs11719165 for leukocytes (p = 0.049, OR = 0.42, 95% CI = 0.17–1.01). However, these are not significant after corrections for multiple testing.
| Risk allele | ADR (CTCAE 3–4) | RAF | Non-ADR (CTCAE 0) | RAF | p-value | OR | 95% CI | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allelic | Dominant | Recessive | Lower | Upper | |||||||||||
| rs11141915 | T | G/G | G/T | T/T | G/G | G/T | T/T | ||||||||
| Platelets | 4 | 21 | 49 | 0.80 | 2 | 18 | 24 | 0.75 | 0.332 | 1.000 | 0.242 | 1.63 | 0.71 | 3.75 | |
| Leukocytes | 1 | 14 | 34 | 0.84 | 3 | 23 | 38 | 0.77 | 0.313 | 0.632 | 0.326 | 0.67 | 0.32 | 1.37 | |
| Neutrophils | 7 | 33 | 56 | 0.76 | 6 | 23 | 44 | 0.76 | 1.000 | 1.000 | 0.875 | 0.92 | 0.47 | 1.79 | |
| Max Leu/Neu | 7 | 34 | 62 | 0.77 | 3 | 20 | 36 | 0.78 | 0.891 | 0.748 | 1.000 | 0.74 | 0.12 | 3.39 | |
| rs1901440 | C | A/A | A/C | C/C | A/A | A/C | C/C | ||||||||
| Platelets | 39 | 26 | 9 | 0.30 | 18 | 23 | 3 | 0.33 | 0.663 | 0.255 | 0.531 | 0.62 | 0.27 | 1.41 | |
| Leukocytes | 22 | 22 | 5 | 0.33 | 22 | 33 | 9 | 0.40 | 0.330 | 0.331 | 0.580 | 0.73 | 0.41 | 1.31 | |
| Neutrophils | 51 | 36 | 9 | 0.28 | 27 | 38 | 8 | 0.37 | 0.099 | 0.043 | 0.799 | 0.52 | 0.26 | 1.01 | |
| Max Leu/Neu | 52 | 42 | 9 | 0.29 | 20 | 32 | 7 | 0.39 | 0.085 | 0.049 | 0.588 | 0.51 | 0.24 | 1.02 | |
| rs12046844 | C | T/T | T/C | C/C | T/T | T/C | C/C | ||||||||
| Platelets | 3 | 23 | 48 | 0.80 | 1 | 14 | 29 | 0.82 | 0.865 | 1.000 | 1.000 | 0.91 | 0.43 | 1.88 | |
| Leukocytes | 1 | 15 | 32 | 0.82 | 1 | 19 | 44 | 0.84 | 0.858 | 1.000 | 0.840 | 0.91 | 0.38 | 2.20 | |
| Neutrophils | 4 | 36 | 56 | 0.77 | 3 | 19 | 51 | 0.83 | 0.221 | 1.000 | 0.148 | 0.61 | 0.30 | 1.20 | |
| Max Leu/Neu | 4 | 36 | 62 | 0.78 | 1 | 17 | 41 | 0.84 | 0.248 | 0.653 | 0.309 | 0.70 | 0.36 | 1.30 | |
| rs11719165 | C | T/T | T/C | C/C | T/T | T/C | C/C | ||||||||
| Platelets | 8 | 39 | 27 | 0.63 | 6 | 18 | 20 | 0.66 | 0.676 | 0.770 | 0.437 | 0.69 | 0.30 | 1.58 | |
| Leukocytes | 4 | 31 | 13 | 0.59 | 5 | 29 | 30 | 0.70 | 0.121 | 1.000 | 0.049 | 0.42 | 0.17 | 1.01 | |
| Neutrophils | 12 | 50 | 34 | 0.61 | 7 | 33 | 33 | 0.68 | 0.253 | 0.629 | 0.208 | 0.67 | 0.34 | 1.30 | |
| Max Leu/Neu | 12 | 53 | 37 | 0.62 | 5 | 28 | 26 | 0.68 | 0.336 | 0.602 | 0.402 | 1.28 | 0.77 | 2.12 | |
- Note: OR and CI are displayed for the test (allelic, dominant or recessive) with the lowest associated p-value, marked in bold, for each toxicity.
- Abbreviations: ADR, adverse drug reaction; CI, confidence interval; CTCAE, Common Terminology Criteria for Adverse Events; OR, odds ratio; RAF, risk allele frequency.
3.4 Toxicity risk scores
All 213 patients were then scored according to their number of homozygous genetic variants. The number of patients for each toxicity risk score was 23 (11%), 64 (30%), 83 (39%), 40 (19%) and 3 (1%), respectively, for scores 0, 1, 2, 3 and 4. It is worth pointing out that, overall, patients with high scores (2–4) were more common in our patient cohort as they accounted for 59% of the patients compared with about 35% in the previous study.14 We also found that three patients were homozygous for all four proposed risk alleles, whereas no such patient was found in the Japanese cohort. Of the three top risk scoring patients, one had a high toxicity profile with CTCAE grades 4 for both platelets and neutrophils (and therefore also for Max Leu/Neu), whereas the other two patients had low toxicity profiles with CTCAE grades 0–1 for all toxicity endpoints.
Based on the risk scores, we then tested if there were any differences in the proportion of risk scores between the patients in the ADR and non-ADR groups using Fisher's exact test, for which the results are listed in Table 6. This showed that there were no significant differences for any of the toxicities on any level as we tested risk scores 2 versus 0–1, 2–4 versus 0–1, 3–4 versus 0–1 and 3–4 versus 0–2.
| Risk score | Platelets | p-value (comparing risk scores) | |
|---|---|---|---|
| ADR (CTCAE 3–4) | Non-ADR (CTCAE 0) | ||
| 0 | 7 | 3 | |
| 1 | 19 | 14 | 1.00 (2 vs. 0–1) |
| 2 | 31 | 20 | 0.84 (2–4 vs. 0–1) |
| 3 | 16 | 6 | 0.44 (3–4 vs. 0–1) |
| 4 | 1 | 1 | 0.48 (3–4 vs. 0–2) |
| Risk score | Leukocytes | p-value (comparing risk scores) | |
|---|---|---|---|
| ADR (CTCAE 3–4) | Non-ADR (CTCAE 0) | ||
| 0 | 5 | 3 | |
| 1 | 15 | 20 | 0.83 (2 vs. 0–1) |
| 2 | 18 | 24 | 0.70 (2–4 vs. 0–1) |
| 3 | 11 | 15 | 0.63 (3–4 vs. 0–1) |
| 4 | 0 | 2 | 0.67 (3–4 vs. 0–2) |
| Risk score | Neutrophils | p-value (comparing risk scores) | |
|---|---|---|---|
| ADR (CTCAE 3–4) | Non-ADR (CTCAE 0) | ||
| 0 | 12 | 3 | |
| 1 | 32 | 22 | 0.17 (2 vs. 0–1) |
| 2 | 34 | 32 | 0.16 (2–4 vs. 0–1) |
| 3 | 17 | 14 | 0.39 (3–4 vs. 0–1) |
| 4 | 1 | 2 | 0.70 (3–4 vs. 0–2) |
| Risk score | Max Leu/Neu | p-value (comparing risk scores) | |
|---|---|---|---|
| ADR (CTCAE 3–4) | Non-ADR (CTCAE 0) | ||
| 0 | 12 | 3 | |
| 1 | 34 | 19 | 0.58 (2 vs. 0–1) |
| 2 | 36 | 22 | 0.41 (2–4 vs. 0–1) |
| 3 | 20 | 13 | 0.39 (3–4 vs. 0–1) |
| 4 | 1 | 2 | 0.56 (3–4 vs. 0–2) |
- Note: p-values from Fisher's exact test comparing the proportions of risk scores.
- Abbreviations: ADR, adverse drug reaction; CTCAE, Common Terminology Criteria for Adverse Events.
4 DISCUSSION
In the present study, we investigated whether four genetic variants, rs11141915, rs1901440, rs12046844 and rs11719165, previously found to be associated with the risk of myelosuppression (specifically leukopenia/neutropenia) in gemcitabine-treated patients14, 15 could be used for risk assessment in our cohort of 213 gemcitabine/carboplatin-treated NSCLC patients. The genotyping showed that the frequencies of the genetic variants in our patients follow the distribution of the alleles in the HapMap CEU population on dbSNP.
The association tests did not show any robust significant associations between the proposed risk alleles and any type of myelosuppressive toxicity in our patients when split into the ADR and non-ADR groups. Two tests were borderline significant, rs1901440 for neutrophils (p = 0.043) and rs11719165 for leukocytes (p = 0.049), although in these cases, the proposed risk alleles trended towards slightly lower levels of toxicity. However, no major conclusions should be drawn based on this as the tests did not withstand adjustments for multiple testing, meaning that the trend of risk alleles towards lower toxicity in our cohort should be interpreted with caution. Similarly, we did not see any significant associations between higher risk scores (combinations of multiple homozygous risk alleles) and myelosuppression. Our study findings suggest that the four genetic variants cannot be applied for the prediction of myelosuppressive ADRs in CEU patients treated with gemcitabine/carboplatin combination chemotherapy. However, the reason for this is probably multifactorial as there are some important differences to highlight between our study and previous studies.14, 15 These considerable differences are related to the treatments, toxicity and study populations. Firstly, the risk alleles and their combinations were overall more common in Swedes than in Japanese patients. Also, the severity of the myelosuppression and the proportion of patients experiencing CTCAE grades 3–4 were higher in our study, which could mask the effects of the genetic variants previously shown, as in the present study 23.0%–48.4% experienced CTCAE grades 3–4, 12.7%–46.9% experienced CTCAE grades 1–2 and 20.7%–34.3% experienced CTCAE grade 0, whereas only 11.3% and 19.2% of patients experienced leukopenia/neutropenia CTCAE grades 3–4, 70.1% and 26.6% experienced CTCAE grades 1–2 and 18.7% and 54.2% experienced CTCAE grade 0 in previous studies,14, 15 respectively. The main difference is that our patients received gemcitabine/carboplatin combination chemotherapy whereas the original study indicated the risk alleles specifically for leukopenia/neutropenia in patients receiving gemcitabine monotherapy. Therefore, the current study might have lost some specificity regarding the risk alleles due to the study set-up. However, as gemcitabine is used in many combinations, it is important to know whether the risk alleles still work as markers of toxicity risk even in combination regimes as well as gemcitabine monotherapy. Unfortunately, the present study indicates that the four investigated genetic markers might not be able to predict myelosuppressive toxicity following gemcitabine/carboplatin combination chemotherapy. Furthermore, as the frequency of the genetic variants and the LD landscape around them differs between the Swedish and Japanese populations, the relationship between the risk alleles and myelosuppression may be lost. Therefore, our results do not mean that the proposed genetic variants are not useful; rather, there is still evidence14, 15 to suggest that the genetic variants can be useful and should be investigated further, mainly in patients treated with gemcitabine monotherapy, although the genetic variant could still be important in combination treatments for other populations, including Japanese patients. Lastly, it may be that the causal genetic variation associated with toxicity is not in LD with the four genetic variants in our Swedish study population.
In conclusion, our results do not indicate any association between the previously identified risk alleles with myelosuppressive ADRs in patients treated with gemcitabine/carboplatin combination chemotherapy. However, the results are important for further understanding of chemotherapy-induced myelosuppression and for pharmacogenetics and the personalised medicine research community working on solving this clinical issue.
ACKNOWLEDGEMENTS
This research was funded by the Swedish Cancer Society (H.G.), the Swedish Research Council (H.G.), Linköping University (H.G.), ALF grants Region Östergötland (H.G.), the Funds of Radiumhemmet (R.L. and L.D.P.) and Marcus Borgströms stiftelse (H.G.). The funders had no role in study design, data collection, data analysis, preparation of the manuscript or decision to publish.
CONFLICT OF INTERESTS
The authors have no conflicts of interest to declare.