Differential decline of SARS-CoV-2-specific antibody levels, innate and adaptive immune cells, and shift of Th1/inflammatory to Th2 serum cytokine levels long after first COVID-19
The authors Bernhard Kratzer and Pia Gattinger equally contributed to the work.
Abstract
Background
SARS-CoV-2 has triggered a pandemic and contributes to long-lasting morbidity. Several studies have investigated immediate cellular and humoral immune responses during acute infection. However, little is known about long-term effects of COVID-19 on the immune system.
Methods
We performed a longitudinal investigation of cellular and humoral immune parameters in 106 non-vaccinated subjects ten weeks (10 w) and ten months (10 m) after their first SARS-CoV-2 infection. Peripheral blood immune cells were analyzed by multiparametric flow cytometry, serum cytokines were examined by multiplex technology. Antibodies specific for the Spike protein (S), the receptor-binding domain (RBD) and the nucleocapsid protein (NC) were determined. All parameters measured 10 w and 10 m after infection were compared with those of a matched, noninfected control group (n = 98).
Results
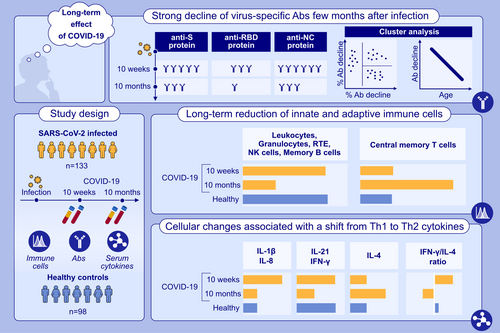
Whole blood flow cytometric analyses revealed that 10 m after COVID-19, convalescent patients compared to controls had reduced absolute granulocyte, monocyte, and lymphocyte counts, involving T, B, and NK cells, in particular CD3+CD45RA+CD62L+CD31+ recent thymic emigrant T cells and non-class-switched CD19+IgD+CD27+ memory B cells. Cellular changes were associated with a reversal from Th1- to Th2-dominated serum cytokine patterns. Strong declines of NC- and S-specific antibody levels were associated with younger age (by 10.3 years, p < .01) and fewer CD3−CD56+ NK and CD19+CD27+ B memory cells. Changes of T-cell subsets at 10 m such as normalization of effector and Treg numbers, decline of RTE, and increase of central memory T cell numbers were independent of antibody decline pattern.
Conclusions
COVID-19 causes long-term reduction of innate and adaptive immune cells which is associated with a Th2 serum cytokine profile. This may provide an immunological mechanism for long-term sequelae after COVID-19.
Graphical Abstract
COVID-19 leads to a sustained reduction of immune cells of the myeloid and lymphoid cell lineages even 10 months after the first infection. Ten months after the first infection, S- and RBD-specific IgG responses declined below the detection limit in almost 18% and in more than 80% in subjects, respectively. Anti-NC antibodies remained positive in all subjects 10 m after the first infection. A shift towards a Th2 cytokine pattern in serum accompanied by an inversion of the IFN-γ/IL-4-ratio was found between the time point 10 weeks and 10 months after infection.Abbreviations: CD, cluster of differentiation; COVID-19, coronavirus disease 2019; IL, interleukin; IFN, interferon; NC, nucleocapsid protein; NK, natural killer; RBD, receptor binding domain; RTE, recent thymic emigrants; S, spike protein; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Abbreviations
-
- ACE2
-
- angiotensin converting enzyme 2
-
- CD
-
- cluster of differentiation
-
- CoV-2
-
- coronavirus 2
-
- COI
-
- cutoff index
-
- COVID-19
-
- coronavirus disease 2019
-
- EBV
-
- Epstein–Barr virus
-
- ELISA
-
- enzyme-linked immunosorbent assay
-
- HC
-
- noninfected control subjects
-
- HLA
-
- human leukocyte antigen
-
- HRP
-
- horseradish peroxidase
-
- HSC
-
- hematopoietic stem cells
-
- Ig
-
- immunoglobulin
-
- NC
-
- nucleocapsid
-
- OD
-
- optical density
-
- PB
-
- peripheral blood
-
- PBMC
-
- peripheral blood mononuclear cells
-
- RBD
-
- receptor-binding domain
-
- RTE
-
- recent thymic emigrant T cells
-
- rtPCR
-
- reverse transcription polymerase chain reaction
-
- S
-
- spike protein
-
- SARS
-
- severe acute respiratory syndrome
-
- TEMRA
-
- T effector memory CD45RA+ T cells
-
- TREC
-
- T cell receptor excision circles
-
- WB
-
- whole blood
-
- 10 m
-
- 10 months after infection
-
- 10 w
-
- 10 weeks after infection
1 INTRODUCTION
SARS-CoV-2 has caused the first Coronavirus pandemic1 and, since 3 years, is responsible for millions of deaths worldwide (https://coronavirus.jhu.edu/map.html). COVID-19 is not only causing acute viral respiratory disease requiring hospitalization in 10%–15% of those infected,2, 3 with intensive care required in 15%–20% of those hospitalized, however,4, 5 COVID-19 also has the unpleasant tendency to cause long-lasting sequelae weeks and even months after acute infection in a subset of 10%–30% of COVID-19 convalescent patients,6, 7 which is also observed after mild acute disease.8 Recently, different risk factors for the development of long-COVID in children have been identified, for example, pre-existing comorbidities and drug treatment, pre-omicron variants, acute phase hospitalization or older age.9 In general, affected patients are commonly referred to as “long-haulers”10, 11 or long-COVID-19 patients.6, 12, 13 As of yet, the exact cause for this clinical picture is ill-defined and mechanistically poorly explored.
However, it is well-known that distinct viral infections have a long-term impact on the immune system and the overall immune homeostasis of those infected and may lead to disease-typical, organ-specific post-viral complications, for example, post-viral bacterial orchitis after mumps virus infection,14 post-measles purulent otitis media or encephalitis,15 but also post-viral bacterial pneumonia (influenza), coagulopathies and fatigue (typical for EBV infections),16 to name just a few. The polymorphic clinical and organ-pathological pattern of long-COVID-19 is, however, unique in its complexity.13 It affects the lungs, causing fibrotic changes with dyspnea, the kidneys, leading to kidney-failure, the cardiovascular system, resulting in heart palpitations and pathological orthostasis as well as the nervous system, leading to tiredness, fatigue, sleep disorders, problems in proper concentration (“brain fog”) but also depressive mood disorders associated with anxiety.17-19
It is tempting to speculate that the quality and quantity of the impact on the immune system of COVID-19 paves the way for such organ-specific pathology, as already observed by the persistence of anti-nuclear antibodies (ANA) in long-COVID-19 patients after resolution of inflammation.20, 21
In a previous study investigating the impact of COVID-19 on the immune system 10 weeks (10 w) after infection, we observed that acute SARS-CoV-2 infection has protracted impacts on the human immune system, even in COVID-19 convalescent patients who underwent a mild disease course.22 In that study, we found a sustained reduction of neutrophil counts which was paralleled by activation of T cells as demonstrated by elevated HLA-DR (CD8+ T cells) and CD38 (CD4+ and CD8+) expression. Moreover, significantly higher numbers of CD3+CD4+CD127+CD45RA− and of CD3+CD8+CD45RO+CCR7− effector memory T cells clearly distinguished COVID-19 convalescent individuals from noninfected control subjects. In addition, numbers of CD19+IgM+CD38+ transitional B cells, as well as plasmablasts (CD19+IgM−CD38+) were higher in convalescents as compared to noninfected controls. Interestingly, 10 w after infection SARS-CoV-2-specific antibody levels were detectable in all but one patient, however, protection as determined by a molecular inhibition assay (MIA), seemed to be variable already at this time point after disease.23, 24
To determine the long-term impact of COVID-19 on the immune system, we re-evaluated the immune status of the previously studied COVID-19 convalescent individuals a second time, 10 months (10 m) after infection. For this purpose, we first determined the antibody levels directed against SARS-CoV-2-specific S-, RBD-, and NC-proteins, and elucidated their RBD-ACE2 blocking capacity as surrogate markers for virus neutralization, respectively. Subsequently, we stratified the COVID-19 convalescent patients according to their antibody waning pattern into three groups followed by multi-parametric flow cytometry-based determination of leukocyte subsets in whole blood with a special focus on T and B lymphocyte subpopulations. In addition, serum cytokine as well as T and NK cellular IFN-γ levels were measured and contrasted to the above-mentioned cellular data, total serum IgE levels, thymic T cell output as determined by T cell receptor excision circles, as well as putative ACE2 expression on recent thymic emigrants (RTE) was measured. We compared the results obtained with an age- and size-matched, noninfected control group, which was recruited in parallel. The control group was negative for SARS-CoV-2 at the time of venipuncture and reportedly asymptomatic 10 w before blood donation.
2 METHODS
2.1 Patients, control subjects and trial conduct
Between May 11, 2020 and August 20, 2020, when SARS-CoV-2 Wuhan Hu-1 was the only circulating virus strain, 133 patients diagnosed with COVID-19 disease were enrolled into this case–control study (Figure 1A). The 133 patients had rtPCR-confirmed (n = 116) and/or SARS-CoV-2 antibody-confirmed (Elecsys® Anti-SARS-CoV-2 assay Roche) (n = 131) COVID-19 disease.25 From these 133 patients, one patient did not show any detectable antibody response against nucleocapsid-, S- or RBD-protein at 10 w and was therefore excluded from further analyses due to the probability of a false positive rtPCR test. Thus, the remaining cohort of 132 patients was analyzed 10 w (77.8 ± 24.6 days) and 10 m (9.5 ± 0.8 months) after infection (Table S1).
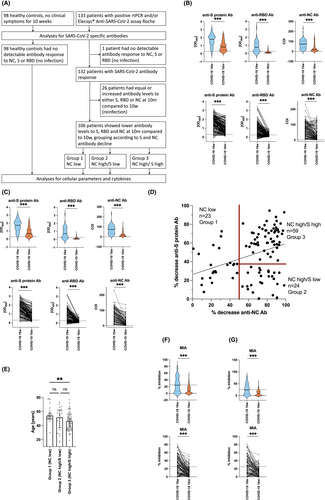
In parallel, 98 noninfected control subjects, who were reportedly asymptomatic for the last 10 w and who were SARS-CoV-2 negative by a certified SARS-CoV-2 antibody test (Elecsys® Anti-SARS-CoV-2 assay Roche) and had a negative rtPCR test for SARS-CoV-2 at the time of venipuncture were enrolled into the study. This cohort is identical to the one published in our previous report on the impact of COVID-19 on the immune system.22 Noninfected control subjects were well-balanced when compared to COVID-19 convalescent patients regarding demographic data, clinical background and drug intake (Table S2) and consisted of 44 males (44.9%) and 54 females (55.1%) with a median age of 51 years (range 14–77) (Table S1).
All individuals gave their written informed consent in accordance with the Declaration of Helsinki. The study was approved by the Ethics Committee of the Medical University of Vienna (EK No: 1302/2020). Venous blood was drawn from all subjects and was either EDTA-anticoagulated (for flow cytometric analyses), heparin-anticoagulated (for cryopreservation of PBMC), or silicon dioxide coagulated (to obtain serum for determining specific antibodies and cytokines).
Since infection-induced antibodies have the principal tendency to drop after an initial peak,26 we have excluded all those COVID-19 convalescent subjects who presented with equal or elevated antibody levels at the end of the observational period of 10 m, to obtain a study population which had experienced only one SARS-CoV-2 infection. This resulted in a study population of 106 patients (Figure 1A). The 106 patients were further divided into three Groups according to their SARS-CoV-2 specific antibody decrease pattern as determined by k-means cluster analyses, which helps to partition a larger number of observations into distinct clusters with minimized within-cluster variances.27 Accordingly, three clusters (Groups) could be assigned. Group 1 subjects were characterized by a low to moderate decrease of NC-specific (≤50%) with variable decreases of S-specific antibody levels; Group 2 showed a high decrease in NC-specific (>50%) but a low decrease in S-specific (≤40%; NC high/S low) antibody levels; Group 3 presented with high decreases of both NC(>50%)- and S(>40%)-specific (NC high/S high) antibody levels (Figure 1A).
2.2 Immunophenotyping by multiparametric flow cytometry
Immunophenotyping was performed by using optimal concentrations of directly conjugated monoclonal antibodies (Table S3A) to leukocyte and lymphocyte (sub) populations according to standard procedures28 as described previously.22 Briefly, whole blood was analyzed freshly on the day of venipuncture (5–9 subjects per day) according to standard, quality-controlled procedures. For a small number of timepoints, some cell populations could not be resolved due to technical reasons, therefore the exact number of patients in the respective analyses is mentioned in each figure.
2.3 Determination of cytokine levels in human serum samples
Cytokines in human serum were determined as described.29 Serum samples from healthy control subjects and COVID-19 patients at 10 w and 10 m were frozen at −80°C directly after sampling and analyses for cytokine levels were performed for all samples at the same time point. A panel of nine different bead regions in two batches with a total of 14 antibody pairs from Thermo Scientific (eBioscience), Biolegend or Miltenyi (Table S3B) were used for the analyses. Beads, 4 × 106 microspheres (Luminex Cooperation, Diasorin, Austin, TX, USA), were coupled with 100 μg capture antibody (one antibody specificity per bead region), as recommended by the manufacturer. For analyses, human serum samples were thawed and centrifuged at 10,000 g for 10 min to remove insoluble precipitates. Subsequently, 30 μL of the serum aliquots were incubated in duplicates with 1.5 × 103 beads per bead region coated with the respective capture antibodies in a total volume of 60 μL at 4°C overnight in the dark on a lab dancer in Multiscreen filter plates (MultiScreen® HTS BV, Merck Millipore, Burlington, MA, USA). On the next day, samples were washed with PBS (Gibco, Thermo Fisher, Waltham, MA, USA), and beads were incubated with 2.5 μg/mL per biotinylated secondary antibodies in a total of 25 μL recognizing the respective cytokines at room temperature for 1 h. Next, another washing step was performed followed by detection of bound antibodies with streptavidin-PE (Biolegend, San Diego, CA, USA) (2 μg/mL) in a total volume of 30 μL. Fluorescence intensities of individual bead populations were determined with a Luminex 100/200 apparatus (Luminex corporation, Austin, TX), related to standard curves obtained using known cytokine concentrations and absolute concentrations were calculated accordingly. The standard curves comprised 13 data points and ranged from 10,000 to 0.0564 pg/mL obtained by three-fold dilution steps.
2.4 Statistical analyses
For the determination of antibody decline the following formula was used: (antibody level 10 w—antibody level 10 m)/(antibody level 10w) × 100. Correlation analyses between the decline of NC- and S-antibody levels revealed three distinct clusters of antibody decline. The k-means algorithm27 was used, to discriminate between similar and dissimilar antibody decline values and to confirm the grouping of each individual accordingly, by setting the expected number of clusters at three.
Prior to statistical evaluation, all metric variables underwent a distribution analysis. It was found that the majority of variables were best fit by a log-normal distribution and consequently log-transformed to obtain a normal distribution. Relative counts of T and B cells were best described by a Johnson type 2 distribution, suggesting a logistic transformation. Data from controls and COVID-19 patients (10 w and 10 m time points) were evaluated by Generalized Estimating Equation (GEE) models with unstructured correlation matrix. This procedure allows analyses of group comparisons and within-subject comparisons of time points in a simultaneous analysis. Groups and time-points were compared by linear contrasts with Sidak sequential correction of p values.
All analyses were performed using Stata 17.0 (StataSoft, College Stations, TX). In general, a p-value below .05 was considered significant. For easier presentation, in graphs we use * denoting p < .05, **denoting p < .01, ***denoting p < .001. Graphs (violin plots, line plots) were prepared by GraphPad 9.5 (GraphPad Software Inc., La Jolla, CA).
Additional material and methods are provided in Appendix A.
3 RESULTS
3.1 Characteristics of COVID-19 convalescent patients with short- and long-term follow up and noninfected control subjects
Here we investigated if there are long-term immunological imprints in a representative COVID-19 study population consisting of 133 individuals by comparing samples obtained 10 w and 10 m after disease onset (Figure 1A). Results obtained for noninfected control subjects were taken as reference. Of note, none of the COVID-19 convalescent patients were vaccinated between the 10 w and 10 m visit, since licensed vaccines became available only later. Furthermore, none of the 133 COVID-19 convalescent patients had reported symptoms of COVID-19 for the period between the two visits. One patient did not show any detectable antibody response against nucleocapsid-, S- or RBD-protein at 10 w and was therefore excluded from further analyses due to the probability of a false positive rtPCR test.
Demographic parameters (Table S1) and COVID-19 unrelated clinical characteristics (Table S2) were similar between the 132 COVID-19 patients and the 98 non-SARS-CoV-2-exposed controls. Of the 132 patients who were infected with the Wuhan Hu-1 virus strain, three patients presented with severe or critical (2.3%), 11 with moderate (8.3%) and 115 with mild (87.1%) disease course, while three remained asymptomatic (2.3%) according to the current WHO classification (https://app.magicapp.org/#/guideline/j1WBYn). The 132 patients were further stratified according to their SARS-CoV-2 serology (Figure 1A). Twenty-six patients were excluded from further analyses due to suspected re-infection, detected by an increase in SARS-CoV-2 specific antibody levels. The remaining 106 patients were stratified according to the observed antibody decline pattern (Figure 1B–D) into three groups (Figure 1A).
With regards to preexisting health conditions, 87 patients (65.9%) in the COVID-19 study group reported comorbidities, among them 68 patients (64.2%) in the antibody decrease group, which was comparable in frequency to 64 individuals in the noninfected control group (65.3%). Among the different comorbidities, allergic diseases in COVID-19 patients (36.4%) and in noninfected controls (43.9%) were the ones with the highest prevalence followed by cardiovascular diseases, metabolic diseases and chronic lung diseases. All other comorbidities (diabetes mellitus, hematopoietic diseases, immunosuppressive conditions, liver, neurological or renal diseases) were present only in few individuals (<10%).
Overall, 60 (45.5%) COVID-19 patients and 48 (49.0%) noninfected control individuals reported regular medication intake to alleviate the above-indicated comorbidities (Table S2). None of the patients reported current or former therapies with lymphoablative biologicals (e.g., rituximab, alemtuzumab, etc.). One patient reported a current cytoreductive treatment, however, this patient was excluded from analyses following the enrolment scheme (Figure 1A). Three individuals received systemic corticosteroids, two in the COVID-19 convalescent and one in the noninfected control group. Therefore, the authors can exclude the influence of medication intake on their data.
3.2 Serum IgG antibody levels specific for S, RBD, and NC decline strongly few months after the first SARS-CoV-2 infection, especially in younger individuals
First, we investigated the levels of SARS-CoV-2-specific IgG antibody levels over the period of 10 m. For this purpose, IgG antibody levels specific for the complete SARS-CoV-2 spike (S) protein, its receptor-binding domain protein (RBD) and the SARS-CoV-2 nucleocapsid protein (NC) were measured. We found that IgG antibodies specific for the latter three antigens were absent in one subject (Figure 1A), which was excluded from the study, and significantly decreased between 10 w and 10 m for the majority of subjects as it is usually expected after an initial peak value after the first infection15 (Figure 1B). However, 26 out of the remaining 132 subjects showed stable or even increased levels of S-, RBD-, or NC-specific antibodies (Figure 1B). Since subjects with such unusually stable or increased virus-specific antibodies may have experienced a clinically silent infection we stratified the COVID-19 convalescent subjects into two subgroups (Figure 1A). The larger group (106 patients) showed decreased (Figure 1C), while the smaller group (26 patients) showed equal or increased antibody reactivity with S-, RBD- or NC-proteins at the second visit at 10 m. The latter group was excluded from further analyses, because it was the goal of the study to investigate the long-term effects of the first SARS-CoV-2 infection in our study population.
Analysis of the remaining 106 COVID-19 convalescent subjects revealed that anti-S protein serum IgG levels significantly decreased from a mean OD405 of 1.651 ± 0.852 at 10 w to 0.853 ± 0.570 at 10 m (p < .0001). Significantly, at 10 w only 3 out of 106 (i.e., 2.8%) COVID-19 convalescent patients had anti-S protein-IgG levels below the cutoff value. This number significantly increased by more than six-fold to 19 out of 106 (17.9%) convalescent patients at 10 m (p = .0005).
The decline of anti-RBD IgG antibody levels, which decreased from a mean OD405-value of 0.825 ± 0.689 at 10 w to 0.166 ± 0.164 (p < .0001) at 10 m was pronounced (Figure 1C). While at 10 w, already 26 out of 106 (24.5%) convalescent patients had anti-RBD serum IgG levels below the cutoff-value, this number more than tripled to 86 out of 106 (81.1%) convalescent subjects at 10 m (p < .0001) (Figure 1C). However, none of the 106 subjects was NC negative, neither at 10 w nor at 10 m, thus, confirming that NC-specific IgG represents the more reliable marker for confirming a previous infection with COVID-19 provided that no NC-containing vaccine30 or inactivated-virus vaccine (Sinovac,31 VLA 200132) was used.
Since RBD-specific antibodies are strongly associated with virus neutralization,23, 24 more than 20% of convalescent subjects 10 w after first infection and more than 80% of convalescent subjects after 10 m of the first infection may have had insufficient levels of SARS-CoV-2-neutralizing antibodies. Indeed, these results were confirmed by a MIA, which serves as surrogate for a virus neutralization test.23, 33 Of the 106 patients with a drop in S-, RBD- and NC-antibodies at 10 w, only 52 patients (49.1%) showed protective neutralizing antibody levels above the cutoff for 25% of inhibition, while at 10 m this number plummeted to 10 patients only (9.4%, p <.0001). The mean percent inhibition dropped from 25.8% at 10 w to 10.8% at 10 m. Only 20 patients showed an inhibition of more than 50% at 10 w, this number dropped to one patient at 10 m.
Likewise, anti-NC antibody levels dropped from a mean cutoff index (COI) value of 72 ± 44 at 10 w to 25 ± 28 (p < .0001) at 10 m (Figure 1C). The percentage decrease in NC antibody levels (Figure 1D) significantly correlated with the decrease in S antibody levels (r = .3461, p = .0003).
The correlation plot of antibody responses appeared to form three distinct patterns of antibody decline (Figure 1D). In fact, k-means algorithm cluster analysis applied to the two-parameter correlation revealed three different groups of patients according to their NC- and S-specific antibody decline pattern. Group 1 had a low decrease in NC (≤50%) and a mixed decrease in S antibody levels (NC low), Group 2 presented with a high decrease in NC (>50%) but a low decrease in S (≤40%; NC high/S low) antibody levels, while Group 3 showed a high decrease in both NC and S (NC high/S high) antibody levels (NC decrease >50%, S decrease >40%) (Figure 1D). Remarkably, the two extreme Groups 1 and 3 significantly differed in their mean age by 10.3 years (56.2 ± 11.5 vs. 45.9 ± 12.3, p = .0072) (Figure 1E). Notably, at 10 w NC antibody levels did not significantly differ between Groups 1 and 3 (86.7 ± 37.4 COI vs. 66.7 ± 45.0 COI; p = .1137) (Figure S1A), while S antibody levels were higher in Group 1 when compared to Group 3 (2.062 ± 0.663 vs. 1.557 ± 0.700 OD405; p = .0089) (Figure S1B). Neutralizing antibody levels similarly dropped in Group 1 (12 [52.2%] vs. 3 [13.0%] patients with inhibition above 25%; p = .0106) and Group 3 (34 [7.6%] vs. 7 [11.9%] respectively, p < .0001). Interestingly, of the patients in Group 2, only 6 (25%) showed an initial blocking activity of more than 25%, while this was not the case in any of the patients at 10 m (Figure S1C).
3.3 COVID-19 impacts on blood leukocyte populations as late as 10 m after infection
Next, leukocyte populations were compared using a 15-parameter flow cytometry approach (Figure 2 and Tables S4A,S4B). In contrast to 10 w, COVID-19 patients had significantly lower total leukocyte counts at 10 m (6.533 ± 1.800 × 106 cells/l vs. 5.749 ± 1.661 × 106 cells/l, p < .0001) and involved neutrophils, monocytes and lymphocytes alike. Lower neutrophil counts in COVID-19 patients compared to noninfected controls were already observed at 10 w, but counts continued to decline until 10 m (4.156 ± 1.419 × 106 cells/l vs. 3.694 ± 1.367 × 106 cells/l, p < .001) (Figure 2). Similar to noninfected control subjects at 10 w, monocyte and lymphocyte counts were found to be lower in COVID-19 convalescent patients at 10 m as compared to 10 w (500 ± 177 × 106 cells/l vs. 452 ± 189 × 106 cells/l, p = .0068; and 1.897 ± 693 × 106 cells/l vs. 1.624 ± 535 × 106 cells/l, p < .0001) (Figure 2, Table S4A). Low lymphocyte counts were equally due to low T, B and NK cell numbers (Figure 2; Table S4A). The described changes were similar for Groups 1–3 for total leucocyte counts and had a similar tendency for total lymphocyte and granulocyte counts, while monocytes were reduced in Groups 2 and 3 but not Group 1 (Figure S2). Reduction of T cells at 10 m affected both the CD3+CD4+ helper and the CD3+CD8+ cytotoxic T cell subsets. Notably, the significant signs of T cell activation observed at 10 w22 affecting both the CD3+CD4+ and CD3+CD8+ T cell subsets and showcased by neo-expression of HLA-DR and CD38,22 largely disappeared at 10 m (Figure 2 and Figure S3A). Figure S3B shows representative dual parameter dot-plots for one control subject and a COVID-19 subject at 10 w and 10 m. Interestingly, at 10 m, the 10 patients within our study who required hospitalization due to severe COVID-19, still presented with elevated numbers of HLA-DR+ and HLA-DR+CD38+ T cells at 10 m, mainly within the CD3+CD8+ subset (Figure S3C) which is in accordance with other studies.34 The described changes in T cell types and subsets thereof were similar for Groups 1–3 (Figure S2), which was clearly different for NK and B cells. In fact, CD56+CD3− NK cell numbers were significantly lower at 10 m (265 ± 139 × 106 cells/l to 216 ± 113 × 106 cells/l, p < .0001) in COVID-19 convalescent subjects when compared to 10 w (Figure 2). However, this drop was mainly attributable to Group 2 (338 ± 220 × 106 cells/l vs. 244 ± 129 × 106 cells/l, p = .0021) and Group 3 (242 ± 99 × 106 cells/l vs. 200 ± 107 × 106 cells/l, p = .0015) (Figure S2A).
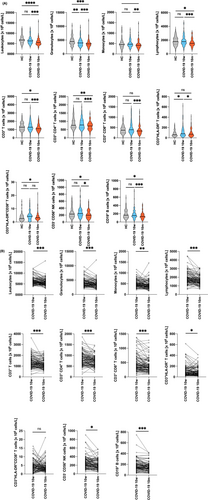
Also, B cell numbers seemed to decrease in the entire COVID-19 study group (Figure 2). However, while they plateaued in Group 1 at 141 ± 81 × 106 cells/l vs. 140 ± 81 × 106 cells/l, p = .8237, they significantly dropped only in Group 2 from 189 ± 142 × 106 cells/l to 148 ± 87 × 106 cells/l, p = .0338 and in Group 3 from 198 ± 92 × 106 cells/l to 161 ± 183 × 106 cells/l, p < .0001 (Figure S2B).
3.4 COVID-19 mainly depletes the pool of RTE T cells but not the central T cell memory as late as 10 m after infection
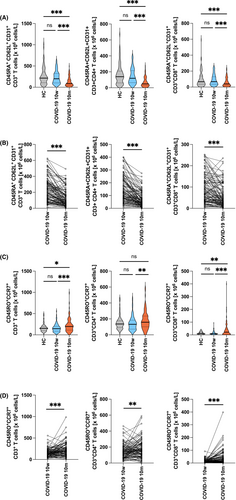
Given the significant reduction of T lymphocyte numbers observed at 10 m, we investigated effects on naïve and memory T cell populations. We found that the most naïve T cells, the RTEs, defined as CD3+CD45RA+CD62L+CD31+ T cells,35 which were similar in numbers to those of the noninfected control subjects at 10 w, were almost cut to half at 10 m (230 ± 150 × 106 cells/l, vs. 126 ± 107 × 106 cells/l, p < .0001), which affected the respective CD4+CD45RA+CD62L+CD31+ helper T cell subset (142 ± 97 × 106 cells/l vs. 68 ± 59 × 106 cells/l; reduction: 52.1%; p < .0001) more drastically than the CD8+CD45RA+CD62L+CD31+ cytotoxic T cell subset (88 ± 66 × 106 cells/l vs. 58 ± 54 × 106 cells/l, reduction: 34.1%; p < .0001) (Figure 3A; Table S5A). The accuracy of flow cytometric RTE determination was self-validated by the fact that patients of younger age still had significantly more circulating RTE numbers when compared to older ones (p < .0001) (Figure S4). Representative results for individuals from different age groups, are shown in Figure S5 and revealed that the COVID-19-dependent drop in RTE numbers at 10 m is observable across all age groups investigated. Significantly, in 93 of 104 (89.4% the CD3+CD4+) and in 83 of 104 (79.8% the CD3+CD8+) of the convalescent patients, the PB RTE subsets were found to be reduced at 10 m when compared to 10 w (Figure 3A,B).
To clarify whether thymic output per se was the reason for the reduced numbers of circulating RTEs in COVID-19 convalescent individuals, TREC analyses on a subset of patient samples were performed. However, we found no evidence for significant reductions of TREC numbers during the observational period (Figure S6).
As already hypothesized above, the significant decrease of circulating RTEs at 10 m was paralleled by an increase of absolute numbers of CD3+CD45RO+CCR7+ central memory CD4+ and CD8+ T cell subsets, that is, from 144 ± 88 × 106 cells/l to 176 ± 113 × 106 cells/l, increase: 22%; p = .0041; and from 15 ± 14 × 106 cells/l, to 39 ± 56 × 106 cells/l, increase: 160%; p = .0001, respectively (Figure 3C,D; and Tables S5A,S5B). Of note, in 55 of 100 (55.0%) convalescent patients the CD4+CD45RO+CCR7+ and in 67 of 98 (68.4%) convalescent patients of the CD8+CD45RO+CCR7+ central memory T cell subsets were found increased at 10 m when compared to 10 w (Figure 3D). Notably, absolute numbers of overall CD45RO+ memory T cells were not increased at 10 m (Figure S7), indicating concurrent contraction of other memory T cell subsets, for example, distinct effector memory cell subsets.
Both, the CD3+CD4+CD127+ effector memory T cell increase and the Foxp3+ CD3+CD4+CD127−CD25+ T regulatory cell decrease observed at 10 w had the tendency to return to almost normal levels at 10 m. (Figure S8; Table S5B). In clear contrast, CD3+CD4+CD45RA+CD127+ cells remained elevated throughout the observational period (140 ± 130 × 106 cells/l and 132 ± 115 × 106 cells/l vs. 79 ± 106 × 106 cells/l, p < .0001 and p < .0001 vs. HC, respectively) (Figure S8; Table S5A) and may reflect expanded CD4+TEMRA (terminal effector memory T cells which re-express CD45RA) cells.36, 37 Interestingly, the changes of T cell subsets were similar in the three Groups of patients with differential antibody decline, with regards to the significant loss of RTE (CD45RA+CD62L+CD31+) and the significant gain (except Group 2) of central memory T cells (CD3+CD45RO+CCR7+) of both CD4+ and CD8+ subsets (Figure S8,S9). In parallel, CD3+CD4+CD127+ effector T cells returned to normal levels in all three groups at 10 m.
3.5 Significant reduction of B cells and B cell subsets between 10 w and 10 m after first COVID-19
Similar to the decline of overall B cell numbers at 10 m (Figure 2), a significant reduction of overall circulating CD19+CD21+CD27+ memory B cells (51 ± 36 × 106 cells/l vs. 39 ± 25 × 106 cells/l, p = .0027) was observed at 10 m compared to 10 w (Figure 4A,D). B memory cell reduction was due to a significant decrease of non-class switched CD19+IgD+CD27+ memory B cells (29 ± 20 × 106 cells/l vs. 23 ± 14 × 106 cells/l, p = .0034) (Figure 4A,D; Table S6). These reductions were paralleled by normalization of CD19+IgM+CD38+ transitional B cell (5 ± 4 × 106 cells/l vs. 8 ± 6 × 106 cells/l, p = .0001) (Figure 4B; Table S6A) and CD19+IgM−CD38+ plasmablast levels (2.7 ± 3.7 × 106 cells/l vs. 1.4 ± 1.1 × 106 cells/l, p = .0121) (Figure 4B and Table S6A) compared to healthy controls. Another interesting finding was the fact that the subset of CD5+ B1-like marginal zone B cells was found to be severely reduced at 10 m in absolute (41 ± 37 × 106 cells/l vs. 24 ± 22 × 106 cells/l, p < .0001) (Figure 4C; Table S6A) and relative (Table S6B) numbers.
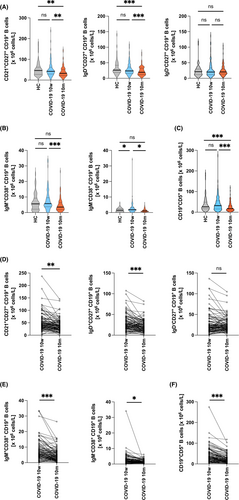
While Groups 2 and 3 presented with significant reductions of non-class switched B cells, a significant reduction of class-switched memory B cells was exclusively seen in Group 3 (Figure S10A–C). In contrast, the drop in CD5+ B cells was observed across all three Groups (Group 1, 34 ± 30 × 106 cells/l to 24 ± 19 × 106 cells/l, p = .0462; Group 2, 43 ± 55 × 106 cells/l to 23 ± 24 × 106 cells/l p < .0001; and Group 3, 42 ± 31 × 106 cells/l to 25 ± 22 × 106 cells/l, p < .0001), respectively (Figure S10D). The normalization of transitional B cell and plasmablast levels was similar among Groups 1–3 (Figure S10E,10F).
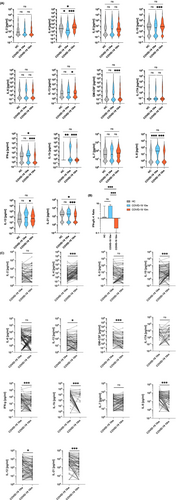
3.6 Systemic serum cytokine levels shift to a type 2-dominated pattern 10 m after COVID-19
Finally, we assessed serum Th1, Th2, Th17 and inflammatory cytokine levels. Figure 5A shows that levels of certain cytokines, which may be considered to be indicative of Th1-dominated inflammation (e.g., IL-1β, IL-8 and IL-12), were higher in sera of COVID-19-convalescent subjects 10 w after infection as compared to the noninfected control group. These alterations resolved 10 m after infection. However, in parallel to this resolution, Th2-dominated serum cytokine patterns emerged, as shown by the significantly elevated IL-4 and IL-10 serum levels (7.3-fold and 1.7-fold, respectively, p < .001 and p = .03, respectively) in sera obtained 10 m as compared to 10 w after infection (Figure 5A). The above-described changes at 10 m were accompanied by significantly lower IFN-γ (p < .0001) and IL-21 (p < .0001) levels compared to noninfected controls, while the overall low IL-17 levels did not change much. Accordingly, the respective IFN-γ/IL-4-ratios of COVID-19 convalescent patients significantly changed from 6.82 to 0.21 between 10 w to 10 m after infection (p < .0001) compared to the balanced ratio of 1.21 as observed in noninfected controls (p = .0486 and p < .0001, respectively). (Figure 5B). No evidence for reduced IFN-γ production on the single cell level was found (Figure S11). Interestingly, Group 1 patients presented with a less pronounced decline in IFN-γ levels, while they showed an increase in IL-10 levels at 10 m (Figure S12).
4 DISCUSSION
We report here on the long-term immune monitoring of 133 patients who had been analyzed 10 w after a first mostly mild infection with Wuhan-strain Hu-1. Our study is rather unique because we were able to perform a follow-up monitoring of immune parameters in the aforementioned patients who did not have another SARS-CoV-2 infection and who were not vaccinated. Thus, we were able to investigate possible long-term effects of a single SARS-CoV-2 infection on the immune system in humans. When comparing the immune parameters measured 10 w and 10 m after the single infection striking differences were found between the two time points. 10 w after COVID-19, patients' had significantly fewer circulating neutrophils compared to noninfected control subjects, while their cytotoxic CD8+ T cells were strongly activated, as reflected by high HLA-DR and CD38 expression levels compared to noninfected controls.22 Overall, these increased numbers of activated HLA-DR+ T cells returned to baseline after resolution of the acute infection while they stayed high in patients who suffered from severe COVID-19. Our findings fit to previous reports,34 and may be a reflection of either antigen persistence, that is, shedding of SARS-CoV-2 particles38, 39 or the more extensive tissue damage in the more severe cases.40, 41 Alternatively, the activated T cells may have been caused by the almost universally increased IL-4 serum levels, found in most of our patients, and the moderately elevated IL-17A levels typically seen in severe cases (p = .031) and as reported previously.42
Moreover, multi-parametric regression analyses had shown that levels of CD3+CD4+ and CD3+CD8+ effector memory cells were elevated, while the levels of CD3+CD25+Foxp3+ T regulatory cells were decreased. In addition, both transitional B cell and plasmablast levels were significantly elevated.22 Such an activation of parts of the immune system at 10 w after infection was confirmed in subsequent studies by others.43-46
Now, the analysis of immune parameters reported in the current study reveals an unexpected change in our study population 10 m after COVID-19. Instead of an activation and expansion of CD3+CD4+ and CD3+CD8+ effector memory cells and transitional B cells and plasmablasts, we found a significant reduction of adaptive immune cells, including T cells (particularly CD3+CD45RA+CD62L+CD31+ RTE) and B cells (non-class-switched CD19+IgD+CD27+ memory B cells) 10 m after COVID-19. Along with the reductions of neutrophils, monocytes and NK cells, it thus seems that a single SARS-CoV-2 infection may cause a long-lasting impact on the cells of the innate and adaptive immune system. Since all leukocyte lineages originate from pluripotent CD34+ hematopoietic stem cells (HSC) in the bone marrow, it is tempting to speculate that such stem cells may have gotten infected by SARS-CoV-2 leading to the herein observed long-term cellular reductions. That CD34+ HSC express ACE2 on the mRNA47 and protein level, and thus display ACE2 enzymatic activity48 has been shown previously. Moreover, evidence exists that CD34+ HSC can be infected by SARS-CoV-2.49 It is therefore quite possible that SARS-CoV-2 infections may target CD34+ HSC directly, explaining the multi-lineage reduction observed after a certain time lag. Alternatively, SARS-CoV-2 infection may have impacted on the bone marrow stromal microenvironment (mesenchymal progenitors, obsteoblasts, fibroblasts and endothelial cells). Recently, it was also suggested that T cells themselves could be the target of an SARS-CoV-2 infection,50 however, we did not find evidence for ACE2 expression on RTE using an ACE2-specific mAb (clone 1.48B) or a RBD-His fusion protein, while both reagents significantly bound to ACE2 transfectants (Figures S13–S16). Thus, it seems more likely that the infection-induced cell damage may indeed occur rather at the stem cell level but further investigations will be necessary to test this hypothesis.
Some more interesting findings were made in our study. Convalescent patients studied 10 m after one SARS-CoV-2 infection presented with significantly up-regulated serum IL-4 levels and a moderate up-regulation of IL-5 was observed. While previous studies have associated eosinopenia with more severe COVID-19 disease,51 the elevated IL-5 serum levels observed herein tend to argue against such a constellation, although eosinophil counts were not directly determined in this study.51 Elevated IL-13 levels observed at 10 w after re-stimulation52 returned to control levels at 10 m (Figure 5). Remarkably, IFN-γ levels, which were comparable to the published literature for the control population and the COVID-19 convalescent subjects at 10 w,53, 54 significantly declined in sera of COVID-19 convalescent subjects at 10 m (Figure 5). Previous studies have shown that patients with severe acute COVID-19 may present with both, either elevated Th1 (IFN-γ, TNF-α, IL-6, IL-2) or Th2 (IL-4 and/or IL-13) cytokine levels.52, 55, 56 These phenotypes were found as early as 2–7 and as late as 14 weeks after infection.56, 57 High IL-4 serum levels may have divergent functions in COVID-19. They may antagonize interferon-driven IgM and IgG3 production,58 lead to reduced FcR-mediated virus defense and correlate with post-acute corona disease.58 On the other hand, high IL-4 levels were shown to inhibit ACE2 expression by lung epithelial cells59 thereby possibly limiting further virus infection of vulnerable tissues during the acute phase of the disease.59 Here we found that IFN-γ levels in sera of COVID-19 convalescent subjects, which at 10 w were similar to noninfected controls,12, 53, 54, 60 significantly decreased at 10 m. The reasons for the drastic decline of serum IFN-γ levels may be found in downregulation of IFN-γ production in individual cells or the mere reduction of NK and/or T cells, both well-known for contributing to serum IFN-γ levels.61, 62 However, we found no evidence that IFN-γ levels were downregulated in single NK or T cells (Figure S11). Thus, the reduced serum IFN-γ levels may be rather the reflection of the general paucity of these two cell types at 10 m. However, also the decline of other IFN-γ producing cell types (e.g., monocytes) may have contributed to the observed phenotype.
While the serum IgE levels significantly correlated with the self-reported allergic status of both the COVID-19 study subjects at 10 w and 10 m (p = .036 and p = .0072, respectively) and the noninfected controls (p = .0094), (Figure S17), no significant changes in serum IgE levels were observed between 10 w and 10 m, in either the allergic or nonallergic subjects, which rather excludes a correlation of the allergic status and the altered IFN-γ/IL-4 ratio. The elevated serum IL-10 levels found at 10 m could be causally linked to the ongoing Treg paucity, and may be capable to increase them.63 However, they may also have resulted from persistent immune activation34 associated with the post-COVID-19 period.64 In that situation, elevated serum IL-4 levels might synergize with IL-10 to suppress overshooting Th1-dominated acute and to potentiate regulatory responses.65 Whether the elevated serum IL-6 levels affected mast cell activation, which has been reported to be associated with post-acute COVID-19 syndrome,66 was not investigated in the present study, as the authors had no possibilities to monitor mast cell numbers and activation status in their patients and controls.
Regarding the relatively well-known decline of SARS-CoV-2-specific antibodies in subjects after a single infection, several additional notable findings were made here. First, we were intrigued by the fact that 10 m after the first infection, S- and RBD-specific IgG antibodies declined below the detection limit in almost 18% and in more than 80% of subjects, respectively. Moreover, more than 90% of patients lacked neutralizing antibody activity at 10 m implying that a large proportion of study subjects after their first infection lost protection from reinfection. Furthermore, we observed a drop in IL-21 levels at 10 m, which might be related to the general decline of T cells at 10 m and may be causally related to the observed decline in antibodies, since IL-21 has been shown to be critical for Tfh cell-mediated B cell expansion and the development of long-lived antibody responses.67
In contrast to S- and RBD-specific antibodies, anti-NC antibodies remained above the detection limit in all subjects 10 m after infection and hence seemed to follow a different kinetic of decline. Whether this was due to differences in antigen persistence38, 39 or different immunogenicity of NC versus S and RBD cannot be answered but may become relevant if NC-targeting SARS-CoV-2 vaccines show clinical benefit.30 Correlation of NC- and S-antibody decline followed by k-means cluster analysis27 revealed another notable result. The antibody decline pattern could cluster the patients in three distinct groups. Group 1 was characterized by a low to moderate decrease in NC specific antibody levels (≤50%), while Group 2 showed a high decrease in NC (>50%) but a low decrease in S (≤40%; NC high/S low) and Group 3 showed a high decrease in both NC and S (NC high/S high) antibody levels (NC decrease >50%, S decrease >40%). Group 1 patients were surprisingly more than 10 years older than Group 3 patients (Figure 1D). The drop in S- and especially RBD-specific antibody levels was confirmed by a Molecular Interaction Assay (MIA).23, 24, 68 Notably, the peculiar antibody constellation in Group 2 was characterized by strong waning of RBD-specific and neutralizing antibodies as detected by MIA, despite the presence of high levels of S-specific antibodies which is indicative of a misdirected antibody response (Figure S1C). In principle, antibody decline was accompanied by a significant shift in the systemic cytokine milieu, which changed from an inflammatory/Th1-driven profile at 10 w, and as described before69 to Th2-dominated immunity at 10 m. However, the magnitude of this shift was group-dependent. Group 1 patients presented with appreciable IFN-γ serum levels at 10 w, while IL-4 levels remained low at this time point and increased only later during the observational period, which led only to a moderate change of the IFN-γ/IL-4-ratio from 1.57 to 0.57 during the observational period, and was paralleled by a significant increase in IL-10 levels, which might act as a survival factor for B cells (Figure S12).70 In contrast, Group 2 and 3 patients presented with IFN-γ-dominated responses at 10 w along with appreciable IL-4 levels, which later during the observational period changed considerably to the opposite, as underlined by the marked changes of IFN-γ/IL-4-ratios from 4.83 to 0.56 and 7.81 to 0.15, respectively. The more dramatic changes of the IFN-γ/IL-4-ratios in Groups 2 and 3 were paralleled by significant decreases in CD19+CD27+ B memory and CD3−CD56+ NK cell numbers (Figure S9A and S3A) while the T cell decline affected all three groups alike. Notably, NK-cells are an important source for systemic IFN-γ levels which may contribute to B memory cell generation. Whether the above described group differences in IFN-γ and NK cell levels are the consequences, for example, of ‘original antigenic sin’71 due to infection with common cold Coronavirus strains HCoV229E,72 HCoV-OC4373 and HCoV-NL63,74 will be the subject of future studies.
Almost all of the COVID-19 convalescent patients who suffered from a post-COVID-19 condition as defined by duration of COVID-19 for more than 28 days according to CDC (https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-care/post-covid-conditions.html) (13 out of 14, 92.9%) had an inverted IFN-γ/IL-4-ratio and were more frequently stratified to Group 3 (NC high/S high antibody decrease, that is, 9 out of 53; 17.0%), compared to Groups 2 (3 out of 24; 12.5%) and 1 (2 out of 23; 8.7%). Notably, the frequency of patients with an inverted IFN-γ/IL-4-ratio was lower in the reminder of the study population (69 out of 92 patients, 75.0%). It is thus tempting to speculate that the inversion of the IFN-γ/IL-4-ratio may contribute to a future collection of biomarkers to identify patients suffering from post-COVID-19 conditions.
This study also has limitations. In fact, our results are mostly based on patients with a mild disease course upon SARS-CoV-2 infection who were able to cure their disease at home and we therefore cannot draw conclusions on how the severity of COVID-19 may be associated with changes in immune parameters. Importantly, the symptoms reported by the study subjects are subjective and we had no possibility to interrogate patients for post COVID-19 syndrome (POCS) emerging during the observational period. Nevertheless, we are not aware of any study which provides evidence for a long-term damage of innate and adaptive immune cells and substantial changes of cytokine parameters in patients after one single SARS-CoV-2 infection. Another limitation of the study is that the recruitment and analyses of the patient samples was performed during the first wave(s) of SARS-CoV-2 infections. Therefore, we here only report on the impact of infections with the first pandemic virus strain, that is, Wuhan Hu-1. However, this weakness may also represent a strength of our study, since it allowed us studying the primary infection in a SARS-CoV-2 naïve patient population and to evaluate the long-term impact of that infection, since due to vaccination and recovery of multiple times, it is currently impossible to investigate such a study collective.
In summary, our results provide a possible explanation that certain manifestations of long-COVID-19 may be associated with damage of the cellular immune system by SARS-CoV-2. This hypothesis can now be investigated in appropriate study populations in detail in the future and this may contribute to the understanding of pathomechanisms underlying long-COVID-19.
AUTHOR CONTRIBUTIONS
B. K., P.G., R. V. and W. F. P. designed research; B. K., P. G., D. T., P. E., U.K., A. R., R. B. S., A. N. A. S., M. F., K. B., I. T., K. G.-P., P. A. T., T. P., I. F., S. W., M.K., G. F. F., R. V., W. F. P., performed research and analyzed data; B. K., P.G., R. V., and W. F. P., wrote the paper. All authors critically read the paper and approved the manuscript. B. K., P.G., R. V. and W. F. P. have verified the underlying data of this study.
ACKNOWLEDGMENTS
We are grateful to Doris Werjant-Locmele and Anna Guentcheva for their help regarding the recruitment and administration of study subjects and to Prof. Pablo Engel (Immunology Unit, Department of Biomedical Sciences, Faculty of Medicine and Medical Sciences, University of Barcelona, Barcelona, Spain) for providing us the ACE2-specific mAb clone 1.48B. We are indebted to all individuals who participated in our study.
FUNDING INFORMATION
This study was supported by grants from the “Medizinisch-Wissenschaftlicher Fonds des Buergermeisters der Bundeshauptstadt Wien” (Medical-Scientific Fund of the Major of Vienna) COVID001 and COVID006; Danube Allergy Research Cluster (Danube ARC) granted by the State of Lower Austria, the Austrian Science Fund (FWF) grant DK-W1248, the Austrian Research Promotion Agency (FFG) COVID-19 emergency call grant 35721032, and Viravaxx, Vienna, Austria. The funders had no role in study design, data collection and analyses, decision to publish or preparation of the manuscript.
CONFLICT OF INTEREST STATEMENT
With regards to the authors disclosure of potential conflicts of interest we would like to indicate that Winfried F. Pickl has received honoraria from Novartis, Astra Zeneca and Roche. Rudolf Valenta has received research grants from HVD Life-Sciences, Vienna, Austria, WORG Pharmaceuticals, Hangzhou, China and from Viravaxx, Vienna, Austria. He serves as consultant for Viravaxx and WORG Pharmaceuticals. The other authors have no conflict of interest to declare. The authors with Russian affiliation declare that they have prepared the article in their “personal capacity” and/or that they are employed at an academic/research institution where research or education is the primary function of the entity.
Open Research
DATA AVAILABILITY STATEMENT
Ethics approval does not allow data sharing of person-related data. Upon reasonable request and depending on a positive ethics vote pseudonymized data can be provided by the principal investigator. All items of the STROBE checklist are covered in this manuscript.