Tonsillar transcriptional profiles in atopic and non-atopic subjects
Abstract
Background
Emerging research suggests that local lymphatic tissue such as tonsils have important role in regulating the immune responses. However, allergen sensitization-induced alterations in transcriptome of tonsils are not known.
Objectives
To examine the key differences in tonsillar gene expression between atopic and non-atopic subjects and further by type of sensitization.
Methods
RNA-sequencing was performed on 52 tonsillar samples from atopic and non-atopic tonsillectomy patients. Sensitization to common food- and aero-allergen was defined by allergen specific IgE. Following groups were studied: (1) aero- and food-allergen sensitized (AS+FS) versus non-sensitized (NS), (2) aeroallergen-sensitized (AS) versus food-allergen sensitized (FS), (3) AS versus NS, (4) FS versus NS. Bioinformatics analysis was done using DESeq2(v3.10.2), WGCNA and GATK pipeline in R software (v3.3.1). Protein–protein interaction network was made from String database.
Results
We studied 13 aeroallergen-sensitized, 6 food-allergen sensitized, 4 both food-and aero-allergen-sensitized and 29 non-sensitized tonsillectomy patients. Overall, 697 unique differentially expressed genes (DEGs) were detected in all sensitized subgroups including chemokines (CXCL2, CXCL8, CXCL10, CXCL11), IL-20RA, MUC1 and MUC20. When comparing different groups, the gene expression profiles overlapped except the AS versus FS group comparison, suggesting significantly different gene expression between the two sensitization subgroups. Furthermore, aeroallergen-sensitized subjects had more prominent immune responses compared with non-sensitized and food-allergen sensitized subjects including gene expression for IL-17 pathway and Toll-like receptor signalling pathway.
Conclusion
Allergic sensitization is associated with extensive tonsillar transcriptomic alterations and changes in immune related genes and pathways. Distinct differences were found between aero-allergen and food-allergen sensitization.
Graphical Abstract
Allergic sensitization, especially to aeroallergens, is extensively associated with tonsillar transcriptomics. Aeroallergen sensitized subjects were characterized by increased expression of cytokines, such as IL-17, and chemokines.Abbreviations: CXCL, C-X-C motif ligand; DEG, differentially regulated genes; IL, interleukin; KEGG, Kyoto Encyclopedia of Genes and Genomes; MUC, mucin; RNA-seq, RNA sequencing
Abbreviations
-
- AS:
-
- Aeroallergen-sensitized
-
- AS+FS:
-
- Aero- and food-allergen sensitized
-
- BP:
-
- Biological process
-
- CC:
-
- Cellular Component
-
- CPM:
-
- Counts per millions
-
- DEGs:
-
- Differentially expressed genes
-
- FDR:
-
- False discovery rate
-
- FS:
-
- Food-allergen sensitized
-
- GO:
-
- Gene Ontology
-
- IgE:
-
- Immunoglobulin E
-
- MF:
-
- Molecular Function
-
- NS:
-
- Non-sensitized
-
- RNA-seq:
-
- RNA-sequencing
-
- TMM:
-
- Trimmed mean of M values
1 INTRODUCTION
The prevalence of allergic sensitization is approximately 30% in developed countries.1 Upper airways serve as the main route of allergens, irritants and pathogens to the body. Hence, interactions between environment, airway barriers and the immune system play an important role in health and pathobiology of allergy.2-5 The palatine tonsils are the secondary nasopharyngeal lymphoid tissue and constitute the first contact point of the immune system to inhaled or ingested pathogens.5 The impaired functioning of tonsils may lead to tonsillar hypertrophy and affect the quality of life especially in children. The most common indications for tonsillectomy in children are recurrent tonsillitis, obstructive sleep disorders and complications of tonsillitis.6, 7
Atopy can affect the tonsillar immune response especially when combined with bacterial or viral infection.7 After contact with antigens, tonsils induce several immune responses where production of pro-inflammatory cytokines and chemokines is integral.8 It has been studied previously that different clinical characteristics and viral infections are associated with distinct immune responses. We have previously shown that tonsillar microbiome also exhibited difference in the atopic compared with non-atopic patients.9 Thus, it is important to study the difference of tonsillar gene expression in atopic compared with non-atopic patients since this has been relatively little studied.
Tonsils provide a unique and important in vivo model for studying the immune responses to microbes and other antigens.9 Although there are many studies related to diseases of palatine tonsils, for example hypertrophy and recurrent tonsillitis; the gene expression in palatine tonsils in atopic and non-atopic subjects is not well studied in detail.10-12 Studying transcriptomic changes of palatine tonsil samples from children with different sensitization patterns will help to understand the lymphoid tissue dynamics in relation to sensitization.
1.1 Aims and hypothesis
The aim was to analyse palatine tonsillar transcriptome to identify key pathways in relation to sensitization pattern by using the RNA-sequencing (RNA-seq). The hypothesis was that there is an association between different sensitization patterns and gene expression profiles of key cytokines.
2 METHODS
The methods for inclusion of study subjects and collection of clinical and laboratory data are shown in Appendix S1. Self-reported allergies and other clinical data were asked with standard questionnaire (Table S2).13 Allergic sensitization was tested with allergen specific immunoglobulin IgE antibodies to any of the common food-allergen (codfish, cow's milk, egg, peanut, wheat, soybean) or aeroallergens (cat, dog, horse, birch, mugwort, timothy, Cladosporium herbarum and Dermatophagoides pteronyssinus) with cut-off level 0.35 kU/L (Phadiatop Combi®, Phadia, Uppsala, Sweden) (Table 1 and Table 2). Animal sensitization was defined as positive IgE antibodies to cat, dog, horse or Dermatophagoides pteronyssinus. Birch, mugwort, timothy and Cladosporium herbarum were considered as pollen aeroallergens.14 Respiratory viruses were detected from nasopharyngeal aspirates and intratonsillar samples by using PCRs (including reverse transcription step when applicable) on nucleic acid extracts.13, 15, 16
| Factor | Atopic (n = 23) | Non-atopic (n = 29) | p-value |
|---|---|---|---|
| Median age (range), years | 12.4 (2.0–38.1) | 7.5 (3.0–21.1) | 0.14 |
| Male | 12 (52%) | 16 (55%) | 0.70 |
| Self-reported allergya | 11 (47%) | 8 (28%) | 0.069 |
| Indication for adeno−/tonsillectomy | |||
| Recurrent tonsillitis | 11 (48%) | 13 (45%) | 0.70 |
| Tonsillar hypertrophy | 11 (48%) | 13 (45%) | 0.94 |
| Other indicationb | 1 (4%) | 3 (10%) | 0.62 |
| Allergen specific sensitization | |||
| Food allergen | 6 (26%) | 0 (0%) | |
| IgE kU/Lc | 1.1 (0.7, 1.7) | ||
| Aeroallergen | 13 (56%) | 0 (0%) | |
| IgE kU/Lc | 4.3 (1.1, 39.1) | ||
| Both food and aeroallergen | 4 (17%) | 0 (0%) | |
| IgE kU/L, foodc | 1.5 (0.6, 10.1) | ||
| IgE kU/L, aeroc | 46.9 (0.6, 98.2) | ||
| Physician diagnosed asthma | 5/21 (27%) | 3/26 (12%) | 0.44 |
| Self-reported allergic rhinitis | 10/19 (53%) | 3/27 (11%) | 0.003 |
| Physician-diagnosed atopic dermatitis | 2/20 (10%) | 6/29 (21%) | 0.32 |
| Upper respiratory symptoms on the operation dayd | 4/18 (22%) | 4/25 (16%) | 0.70 |
| Cough on the operation day | 2/18 (4%) | 1/25 (11%) | 0.56 |
| Respiratory viruses in tonsillar tissue | 11 (48%) | 15 (52%) | 0.78 |
| Respiratory viruses in nasopharynx | 18 (75%) | 24 (83%) | 0.49 |
| Smoking or exposure to smoking | 10/22 (45%) | 15/28 (54%) | 0.57 |
| Use of antibiotics within 1 year | 15/19 (79%) | 18/28 (64%) | 0.28 |
| Season of sampling | |||
| April–August (in-season) | 9 (39%) | 11 (38%) | 0.93 |
| September–March (off-season) | 14 (61%) | 18 (62%) | 0.93 |
- Note: Data are expressed as number of subjects (%) except age. Differences between atopic vs. nonatopic subjects were calculated with Mann–Whitney U test, Chi square test or Fischer exact test (when counts <5). Allergen specific sensitization was defined as positive immunoglobulin (Ig) E antibody (≥0.35 kU/L) to any of the following allergens: codfish, cow's milk, egg, peanut, soybean, wheat, cat, dog, horse, birch, mugwort, timothy, Cladosporium herbarum and Dermatophagoides pteronyssinus (Phadiatop Combi®, Phadia, Uppsala, Sweden). The latter eight allergens were considered as aeroallergens.
- a Animal or housedust mite, pollen, food or drug allergy.
- b Chronic white patches in tonsils (n = 1), teeth braces (n = 1), periodic fever (n = 2).
- c Data expressed as median (IQR).
- d Two had rhinitis, two had throat pain, two had cough and rhinitis, one had cough and throat pain and one had rhinitis and throat pain.
| Factor | Aeroallergen sensitized (AS) n = 13 | Food allergen sensitized (FS) n = 6 | Aero- and food-allergen sensitized (A + FS) n = 4 | Non-sensitized (NS) n = 29 | p-value |
|---|---|---|---|---|---|
| Median age (range), years | 14.2 (7.5–38.1) | 4.6 (2.0–15.5) | 6.8 (4.1–18.5) | 7.5 (3.0–21.1) | 0.006 |
| Male | 8 (62%) | 2 (33%) | 2 (50%) | 16 (55%) | 0.71 |
| Self-reported allergya | 6/11 (55%) | 2/5 (40%) | 3 (75%) | 8/26 (31%) | 0.28 |
| Indication for adeno−/tonsillectomy | |||||
| Recurrent tonsillitis | 9 (69%) | 1 (17%) | 1 (25%) | 13 (45%) | 0.13 |
| Tonsillar hypertrophy | 4 (31%) | 5 (83%) | 2 (50%) | 13 (45%) | 0.20 |
| Other indicationb | 0 | 0 | 1 (25%) | 3 (10%) | 0.31 |
| Physician diagnosed asthma | 2/12 (17%) | 2/5 (40%) | 1 (25%) | 3/26 (12%) | 0.46 |
| Self-reported allergic rhinitis | 6/10 (60%) | 1/5 (20%) | 3 (75%) | 3/27 (11%) | 0.004 |
| Physician-diagnosed atopic dermatitis | 0 | 0 | 2 (50%) | 6 (21%) | 0.088 |
| Upper respiratory symptoms on the operation dayc | 2/9 (22%) | 1/5 (20%) | 1 (25%) | 4/25 (16%) | 0.96 |
| Cough on the operation day | 1/5 (20%) | 1/9 (11%) | 0 | 1/25 (4%) | 0.54 |
| Respiratory viruses in tonsillar tissue | 7 (54%) | 2 (33%) | 2 (50%) | 15 (52%) | 0.86 |
| Respiratory viruses in nasopharynx | 9 (69%) | 6 (100%) | 3 (75%) | 24 (83%) | 0.44 |
| Smoking or exposure to smoking | 7/12 (58%) | 3 (50%) | 0 | 15/28 (54%) | 0.21 |
| Use of antibiotics within 1 year | 8/10 (80%) | 5/5 (100%) | 2/4 (50%) | 18/28 (64%) | 0.28 |
| Season of sampling | |||||
| April–August | 4 (31%) | 2 (33%) | 3 (75%) | 11 (38%) | 0.45 |
| September–March | 9 (69%) | 4 (67%) | 1 (25%) | 18 (62%) | 0.45 |
- Note: Data are expressed as number of subjects (%) except age. Differences between the groups were calculated with Kruskal–Wallis test, Chi square test or Fischer exact test (when counts <5). Allergen specific sensitization was defined as positive immunoglobulin (Ig) E antibody (≥0.35 kU/L) to any of the following allergens: codfish, cow's milk, egg, peanut, soybean, wheat, cat, dog, horse, birch, mugwort, timothy, Cladosporium herbarum and Dermatophagoides pteronyssinus (Phadiatop Combi®, Phadia, Uppsala, Sweden). The latter eight allergens were considered as aeroallergens.
- a Animal or housedust mite, pollen, food or drug allergy.
- b Chronic white patches in tonsils (n = 1), teeth braces (n = 1), periodic fever (n = 2).
- c Two had rhinitis, two had throat pain, two had cough and rhinitis, one had cough and throat pain and one had rhinitis and throat pain.
2.1 Definitions
- Atopy (sensitization) was defined as positive immunoglobulin (Ig) E antibody (≥0.35 kU/L) to any of the following allergens: codfish, cow's milk, egg, peanut, soybean, wheat, cat, dog, horse, birch, mugwort, timothy, Cladosporium herbarum and Dermatophagoides pteronyssinus (Phadiatop Combi®, Phadia, Uppsala, Sweden).
- Food-allergen sensitization was defined as positive IgE antibodies to any of the former 6 allergens.
- Aero-allergen sensitization was defined as positive IgE antibodies to any of the latter 8 allergens.
- Highly sensitized was defined as IgE > 10 kU/L to any of the allergens detected.
- Allergic symptoms were defined as self-reported allergic rhinitis, doctor-diagnosed atopic eczema (asked in the questionnaire, Table S2) and doctor-diagnosed asthma (asked in the questionnaire).
- Virus infection was defined by positive virus PCR test from nasopharyngeal aspirate (NPA) or tonsil, and virus coinfection if two or more virus PCR tests were positive.
- Tonsillar hypertrophy was defined as enlarged tonsils causing symptoms, for example obstructive sleep disorder, snoring and eating problems.
- Recurrent tonsillitis was defined as several pharyngitis in the medical records.
2.2 Comparisons
Tonsillar whole-transcriptome gene expression was compared between the following predefined groups:
Atopic (allergic sensitized) versus non-atopic (non-sensitized).
2.3 Subgroup's analysis
1. Aero- and food-allergen sensitized (AS+FS) versus non-sensitized (NS).
2. Aeroallergen-sensitized (AS) versus food-sensitized (FS).
3. Aeroallergen-sensitized (AS) versus non-sensitized (NS).
4. Food-sensitized (FS) versus non-sensitized (NS).
2.4 Additional, exploratory comparisons
5. Highly sensitized (allergen specific IgE sensitization >10 kU/L) versus non-sensitized.
6. Allergic + sensitized versus non-allergic-non-sensitized.
2.5 RNA-sequencing, data processing and read mapping
Total RNA from cell samples was isolated using the RNeasy mini kit (Qiagen, Hilden, Germany). Reverse transcription was performed with the Revert Aid M-MuLV Reverse Transcriptase (Fermentas, St. Leon-Rot, Germany) using random hexamer primers according to the manufacturers. RNA-sequencing on the samples was carried out with Illumina HiSeq 2500. Preprocessing and quality control of reads were performed with FastQC (v0.10.1)17 and the filtered single-end reads were aligned to the human genome (GRch38) using HISAT (v2.1.0).18 The number of uniquely mapped reads varied between 41 and 50 million per sample.
2.6 Bioinformatics analysis of RNA-sequencing data
Raw read counts were transformed into normalized data using the counts function in Deseq219 R software (v3.3.1). Differentially expressed genes (DEGs) were identified with Deseq2 (v3.10.2) threshold for statistical significance was set at a false discovery rate (FDR) < 0.05 and log2 (foldchange) greater than 1 or less than −1. Hierarchical clustering of genes was done using Pearson correlation and ward linkage. A heatmap of gene expression matrix was produced with Pheatmap R package (v3.3.1). The weighted gene coexpression network analysis20 algorithm was used to construct networks for gene expression from all study participants.
2.7 Gene ontology analysis and functional protein network analysis
Gene ontology (GO) and pathway enrichment analysis was performed with cluster profiler.21 Functional protein association networks were investigated by using STRING (http://string-db.org/). Cytokines clustering analysis was performed by using in-house collected clusters of cytokines.
2.8 Variants analysis
In order to find the regulatory variants responsible for the differential expression, we performed eQTL analysis which identifies cis and trans regulatory elements. For this purpose, variant calling from the HISat2 alignments was performed using Genome Analysis Toolkit (GATK)22 following the best practices (https://gatk.broadinstitute.org/hc/en-us/articles/360035531192-RNAseq-short-variant-discovery-SNPs-Indels-; last accessed 03.08.2020). The SNPs were functionally annotated using ANNOVAR.23 The correlation between the gene expression and the expressed SNPs was assessed using ReQTL implemented in R.24 Finally, SNPs that passed the filtering criteria were subjected to eQTL analysis (p value cis < 1e-6; p value trans <1e-6) along-with the batch corrected gene expression matrix, sensitization pattern and other covariates (Age groups and sex).
3 RESULTS
3.1 Differential gene expression in palatine tonsil samples
3.1.1 Atopic versus non-atopic
To obtain an overview of the tonsillar gene expression differences in atopic (n = 23) and non-atopic (n = 29) subjects, we compared the RNA-seq data in these groups. A linear regression model was fitted to the data to analyse the variation at the group level. However, the comparison of atopic and non-atopic subjects showed no statistically significant results (FDR <0.05). Most interesting findings were found from subgroup comparisons (see below).
3.1.2 Aero- and food-allergen sensitization versus no sensitization
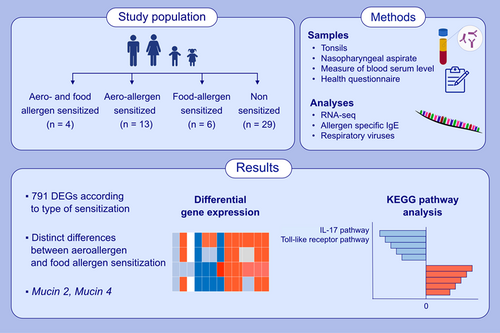
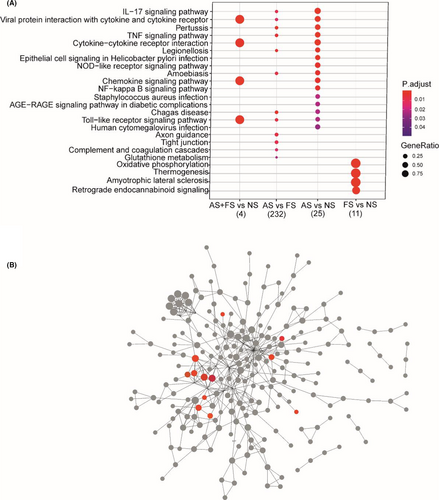
When comparing AS+FS and NS groups, a total of 97 (5 up- and 92 down-regulated) DEGs were identified (FDR <0.05 and log2 (foldchange) greater than 1 or less than −1) (Figure 1A, Table E1 in this article's Online Repository). Of these 97 DEGs, the top upregulated genes were CXCL10, CXCL11 and CXCL9 (Table 3). DEGs in this group were found to be enriched in KEGG pathways viral protein interaction with cytokine and cytokine receptor, cytokine–cytokine receptor interaction, chemokine signalling pathway and Toll-like receptor signalling pathway (Figure 2A). The downregulated genes were enriched in GO BP cornification, keratinization and cornified envelope (Table GO results in this article's Online Repository).
| Rank | Genes | Description | Function | Fold Change | Log2FC | FDR | Comment (references) |
|---|---|---|---|---|---|---|---|
| Immune related DEGs in AS+FS versus NS subjects | |||||||
| 1. | CXCL9 | C-X-C motif chemokine Ligand 9 | This antimicrobial gene encodes secreted proteins involved in immunoregulatory and inflammatory processes. | 2.25 | 1.17 | 0.01 | |
| 2. | IP-10 (CXCL10) | C-X-C motif chemokine Ligand 10 | Stimulation of monocytes, natural killer and T-cell migration | 4.08 | 2.03 | 0.001 | Allergic inflammation,25 Asthmatic airway26 |
| 3. | CXCL11 | C-X-C motif chemokine Ligand 11 | Development, homeostasis and function of the immune system | 3.91 | 1.97 | 0.005 | Allergic inflammation25 |
| Immune related DEGs in AS versus FS subjects | |||||||
| 1. | CXCL2 | C-X-C motif chemokine Ligand 2 |
Antimicrobial gene involved in immunoregulatory and inflammatory processes |
2.5 | 1.3 | 0.04 | |
| 2. | CXCL3 | C-X-C motif chemokine Ligand 3 | Plays a role in inflammation and as a chemoattractant for neutrophils | 3.6 | 1.85 | 0.04 | |
| 3. | CXCL5 | C-X-C motif chemokine Ligand 5 | Involved in recruitment of leukocytes and neutrophils | 6.6 | 0.002 | ||
| 4. | CXCL8 | C-X-C motif chemokine Ligand 8 (also known as IL-8) | Major mediator of the inflammatory response | 0.25 | 2.11 | 0.03 | |
| 5. | CXCL9 | C-X-C motif chemokine Ligand 9 | 5.76 | 2.4 | 0.02 | ||
| 6. | IP-10 (CXCL10) | C-X-C motif chemokine Ligand 10 | Stimulation of monocytes, natural killer and T-cell migration | 4.84 | 2.2 | 0.002 | Allergic inflammation,25 Asthmatic airway26 |
| 7. | CXCL11 | C-X-C motif chemokine Ligand 11 | Development, homeostasis, and function of the immune system | 5.6 | 2.5 | 0.005 | Allergic inflammation25 |
| 8. | IL20RA | Interleukin 20 receptor subunit alpha | May be involved in epidermal function | 2.8 | 1.5 | 0.01 | |
| 9. | MUC1 | Mucin 1, Cell Surface Associated | Forms protective mucous barriers on epithelial surfaces | 5.6 | 2.5 | 0.02 | |
| 10. | MUC4 | Mucin 4, Cell Surface Associated | Plays important roles in the protection of the epithelial cells and have been implicated in epithelial renewal and differentiation | 5.2 | 2.4 | 0.04 | |
| 11. | MUC20 | Mucin 20, Cell Surface Associated | Mucous barrier | 3.5 | 1.8 | 0.01 | |
| 12. | DEFB4A | Defensin Beta 4A | Antibiotic peptide which is locally regulated by inflammation | 10.5 | 3.4 | 0.03 | |
| 13. | DEFB1 | Defensin Beta 1 | Antimicrobial peptide implicated in the resistance of epithelial surfaces to microbial colonization | 3.24 | 1.7 | 0.04 | |
| Immune related DEGs in AS versus NS subjects | |||||||
| 1. | VSIG2 | V-Set and Immunoglobulin Domain Containing 2 | Predicted to be integral component of plasma membrane | 2.2 | 1.13 | 0.04 | |
| 2. | LGALS7 | Lectin, Galactoside-Binding, Soluble, 7 | Carbohydrate binding | 4.02 | 2.01 | 0.01 | |
| 3. | FOS | Fos Proto-Oncogene | Regulators of cell proliferation, differentiation, and transformation | 2.6 | 1.41 | 0.01 | |
| 4. | DEFB4A | Defensin Beta 4A | Antibiotic peptide which is locally regulated by inflammation | 2.75 | 1.46 | 0.03 | |
| 5. | CXCL2 | C-X-C motif chemokine Ligand 2 |
Expressed at sites of inflammation, antimicrobial gene involved in immunoregulatory and inflammatory processes |
2.26 | 1.18 | 0.03 |
Related to asthma27 |
| 6. | CXCL3 | C-X-C motif chemokine Ligand 3 | Plays a role in inflammation and as a chemoattractant for neutrophils | 2.46 | 1.30 | 0.047 | |
| 7. | CXCL5 | C-X-C motif chemokine Ligand 5 | Involved in the recruitment and activation of leukocytes | 18.37 | 4.20 | 0.002 | |
| 8. | CXCL8 | C-X-C motif chemokine Ligand 8 (also known as IL-8) | Major mediator of the inflammatory response | 2.49 | 1.32 | 0.03 | |
| 9. | IL1B | Interleukin-1 B | An important mediator of the inflammatory response | 2.28 | 1.19 | 0.03 | |
| 7. | CXCR2 | C-X-C Motif Chemokine Receptor 2 (also known as IL8RA) | Receptor for interleukin-8 which is a powerful neutrophil chemotactic factor | 2.5 | 1.31 | 0.01 | |
| 9. | MUC1 | Mucin 1, Cell Surface Associated | Forms protective mucous barriers on epithelial surfaces | 2.08 | 1.06 | 0.02 | Suppresses response to bacterial infection27 |
| 10. | MUC20 | Mucin 20, Cell Surface Associated | Mucous barrier | 2.07 | 1.05 | 0.01 | Related to asthma28 |
| 11. | IL-20RA | Interleukin-20 Receptor Subunit Alpha | A cytokine that may be involved in epidermal function | 2.04 | 1.03 | 0.01 | |
| Immune-related DEGs in FS versus NS subjects | |||||||
| 1. | CXCL11 | C-X-C motif chemokine Ligand 11 | Development, homeostasis, and function of the immune system | 0.20 | −2.26 | 0.005 | Allergic inflammation25 |
| 2. | IP-10 (CXCL10) | C-X-C motif chemokine Ligand 10 | Stimulation of monocytes, natural killer and T-cell migration | 0.35 | −2.07 | 0.002 | Allergic inflammation25 Asthmatic airway26 |
- Note: References correspond to the phenotype that has been studied before in relation to the susceptible gene. Foldchanges refer to the first group compared with the second group, for example foldchange of 1 would mean gene is upregulated in the first group.
3.1.3 Aeroallergen sensitization versus food-allergen sensitization
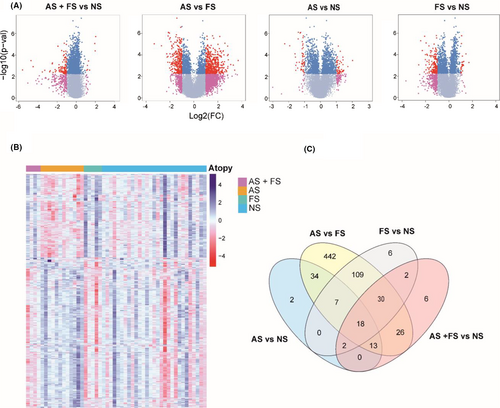
When comparing AS and FS groups, a total of 680 (495 up- and 185 down-regulated) DEGs were identified (FDR <0.05 and log2 (foldchange) greater than 1 or less than −1) (Figure 1A, Table E2 in this article's Online Repository). Of these 680 DEGs, some DEGs were linked to immune response, for example CXCL2, CXCL3, CXCL5, CXCL8, CXCL9, CXCL10, CXCL11, CXCL2, MUC1, MUC4, MUC20, DEFB4A and DEFB1 (Table 3). Upregulated genes in AS subjects were enriched in GO BP including inflammatory response, cytokine mediated signalling pathway, neutrophil chemotaxis and neutrophil degranulation. Upregulated genes were also enriched for KEGG pathways such as Pertussis, IL-17 signalling pathway, viral protein interaction with cytokine and cytokine receptor and Toll-like receptor signalling pathway (Figure 2A). KEGG enrichment for downregulated genes was found for oxidative phosphorylation and Fanconi anaemia pathway (Table GO results in this article's Online Repository).
3.1.4 Aeroallergen sensitization versus no sensitization
When comparing AS and NS groups, a total of 76 (56 up- and 20 down-regulated) DEGs were identified (FDR <0.05 and log2 (foldchange) greater than 1 or less than −1) (Figure 1A, Table E3 in this article's Online Repository). We found several interesting, upregulated genes in the comparison AS versus NS group including VSIG2, IL-20RA, LGALS7, DEFB4A, FOS, CXCL2, CXCL3, CXCL5, CXCL8, DEFB1 and CXCR2 (Table 3). KEGG pathway enrichment showed the upregulation of genes for IL-17 pathway, viral protein interaction with cytokine and cytokine receptor, cytokine–cytokine receptor interaction, chemokine signalling pathway and Toll-like receptor signalling pathway, NF-Kappa B signalling pathway and NOD-like receptor signalling pathway (Figure 2A).
The upregulated genes in this comparison and from other comparisons were clustered into a fine string network (Figure 2B). These upregulated genes were found to be enriched in chemokine-mediated signalling pathway, cytokine-mediated signalling pathway, neutrophil chemotaxis, inflammatory response and response to bacterium. There were no enrichment results found for the downregulated genes in this group.
3.1.5 Food-allergen sensitization versus no sensitization
When comparing FS and NS groups, a total of 174 (14 up- and 160 down-regulated) DEGs were identified (FDR <0.05 and log2 (foldchange) greater than 1 or less than −1) (Figure 1A, Table E4 in this article's Online Repository) These DEGs included CXCL5, CXCL9, CXCL10 and CXCL11 (Table 3). Upregulated genes were enriched in several GO BP including Electron transport coupled proton transport, oxidative phosphorylation and aerobic respiration (Figure 2A & B). Downregulated genes were enriched in GO BP Cornification, epithelial cell differentiation and programmed cell death.
3.2 Overview of tonsillar RNA-sequencing transcriptome
Pairwise comparisons (Figure 1A) showed different levels of differentially expressed genes with significant differences in AS versus FS group (Table E2). Many DEGs were observed overlapping in all group comparisons except the AS versus FS group that had 442 different DEGs (Figure 1C), suggesting that the gene expression is significantly different in these two types of sensitizations. The overall unique DEGs in all group comparisons are shown in heatmap (Figure 1B), with 97, 680, 76 and 174 DEGs in AS+FS versus NS, AS versus FS, AS versus NS and FS versus NS respectively.
Interestingly, AS+FS versus NS and FS versus NS exhibited same overall expression profile while AS versus NS and AS versus FS showed different pattern of gene expression. Mainly, DEGs in AS subjects showed a distinct expression profile in the heatmap. In addition, immune-related gene cluster analysis showed distinct gene expression in all group comparisons (Figure 3A,B).
3.3 Network analysis
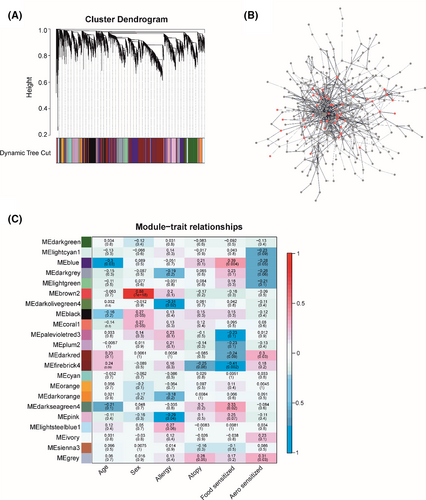
We constructed weighted gene co-expression networks of the RNA-seq data to perform functional classification related to each subject group. WGCNA (Weighted Gene Co-expression Network Analysis) analysis can identify clusters or modules of genes that have similar gene expression pattern. Hereby, we hypothesize that modules of genes that are co-expressed are most likely to participate in same biological functions. In this analysis, we identified 22 modules (Figure 4A,C); 5 modules were found statistically significant in self-reported allergy, food and aeroallergen sensitization. Blue module (Figure 4C) was positively associated with food-allergen sensitization and negatively associated with aeroallergen sensitization. Functional enrichment of genes in blue module showed enrichment in Fanconi anaemia pathway, B-cell receptor signalling pathway and viral carcinogenesis. This functional characterization suggests that genes co-expressed in blue module is related to atopy. GO BP (Biological Process) enrichment showed results in viral process. Pink module that was enriched in allergy, the genes in this module were enriched in antigen processing: Ubiquitination and Proteasome degradation and ISG1-antiviral mechanism. We identified significantly overrepresented biological pathways and gene ontology terms for each module in the network. GO analysis revealed the enrichment in response to organic substance and viral process. Protein–protein interaction network of genes clustered in firebrick4 module showed the interaction of the proteins (Figure 4B). GO enrichment of genes in red colour nodes exhibited response to cytokine stimulus, regulation of inflammatory response, regulation of cytokine production and viral process.
3.4 Variants analysis
We performed variant analysis of RNA-seq data to identify the genomic variants that were associated with sensitization and types of sensitizations. Altogether, 56,872 filtered SNPs were employed for the eQTL analysis. Of these, a total of 640 susceptibility regulatory variants (2 cis- and 638 trans) were having an FDR value of <0.05, indicating the most robust changes that were associated with sensitization. Our results showed that these susceptibility variants can affect the expression of 109 DEGs in tonsillar RNA-seq data (see variants data table in this article's Online Repository). The results suggested significant SNPs including cis regulators rs1049092 (missense variant) and rs118027159 (intron variant), for HLA-DQB1 and TUG1 respectively. Other clinically interesting variants included 116 trans SNPs that were significantly associated with the expression of IL-1B gene. (Table 4, see variants data table in this article's Online Repository). Furthermore, we identified novel trans variants for MAPK3, MUC-21, IL-1B and HLA-DQB1. The variants with the known association with the phenotype and genes with large number of variants are shown in Table 4.
| Ensembl gene id | Regulation | Number of SNPs | Mapped gene | Location | Related Trait (s) GWAS catalogue | Beta | t-stat | p-value | FDR |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000179344 | cis | 1 | HLA-DQB1 | 6:32662025_A>G | Asthma, allergic sensitization | −1.383 | −8.081 | 1.24E-10 | 9.47E-06 |
| ENSG00000099985 | cis | 1 | OSM | 22:30974057_A>G | 6.268 | 5.748 | 5.37E-07 | 0.00583 | |
| ENSG00000125538 | trans | 116 | IL-1B | (see SNPs table) | |||||
| ENSG00000204544 | trans | 1 | MUC21 | 11:1956322_G>A | Asthma (childhood onset) | −6.222 | −5.660 | 7.33E-07 | 0.03 |
| ENSG00000102882 | trans | 1 | MAPK3 | 12:49269847_T>C | Tonsillectomy | −1.910 | −5.764 | 5.08E-07 | 0.03 |
| ENSG00000173391 | trans | 112 | OLR1 | (see SNPs table) | |||||
| ENSG00000105963 | trans | 100 | ADAP1 | ||||||
| ENSG00000081041 | trans | 53 | CXCL2 | ||||||
| ENSG00000099985 | trans | 50 | OSM |
- Note: References correspond to the phenotype that has been studied before in relation to the susceptible gene.
4 DISCUSSION
This is the first study to evaluate palatine tonsillar RNA-seq transcriptomic alterations in relation to sensitization patterns. Our data provide new insights into the gene expression of tonsils. No overall DEGs were found between atopic and non-atopic groups. However, large number of DEGs were found in comparing sensitization subgroups. As expected, many subgroup comparisons showed that specific sensitization was associated with overrepresented chemokines (CXCL2, CXCL8, CXCL10 and CXCL11) and genes from MUC-family (MUC1, MUC20). Interestingly, aeroallergen versus food sensitized subjects showed 63% distinctively different genes. Furthermore, aeroallergen sensitized subjects had more prominent immune responses including gene expression for IL-17 pathway. We are also first to investigate the effect and association of gene variants with the allergic sensitization across the tonsillar transcriptome.
The exploration of the DEGs identified in AS+FS versus NS group found that in addition to other highly upregulated genes, especially the chemokines (CXCL10 and CXCL11) showed elevated expression. These are proinflammatory chemokines that regulate and maintain inflammatory immune responses by mobilizing the immune cells to the infection site. CXCL10 can worsen allergic airway inflammation.29 In addition, CXCL10 has recently been found to be associated with food allergy in very young children.30 AS versus NS subjects on the other hand, showed the upregulation of chemokines (CXCL2, CXCL8/IL-8) which are involved in regulating immune responses in inflammatory diseases.27, 31-33 IL-8 is a major inflammatory mediator31 known to be stimulated by environmental changes, bacterial infection or IL-1 gene expression.34 IL-8 has been found in airway epithelial cells upon the exposure of allergens and may act as chemoattractant in allergic individuals.35 IL-8 binds to the G-protein couple receptors CXCR1 and CXCR2 for inducing the inflammatory response.32 Interestingly, we also found the upregulation of IL-8 receptor, that is CXCR2 in AS subjects. IL-8 is mainly involved in inducing the inflammatory response by recruiting the neutrophils, mononuclear phagocytes, mast cells and T cells. IL-8 contributes to the pathogenesis of several respiratory diseases including bronchial asthma, acute respiratory distress syndrome, respiratory syncytial virus infections and viral bronchiolitis.32, 33, 36 Moreover, IL-8 may play crucial role in the onset of acute asthma exacerbations or severity of allergic asthma.1, 2 Therefore, it is implicated to control the atopic asthma by mediating the IgE production.37 Since IL-8 and CXCL2 are important part of inflammatory processes, their aberrant expression can also mediate the inflammatory processes in tonsils of aero-sensitized subjects.
In addition, DEGs from MUC family (MUC1, MUC20) were upregulated in AS versus NS and AS versus FS subjects. MUC1 is an anti-inflammatory gene of mucin family expressed in nasal, pharyngeal, bronchial and tracheal cells in the human airway and in lung epithelial cells.38, 39 MUC1 is known for its function in many inflammatory diseases. For example, Li-Bo et al40 reported significantly decreased expression of MUC1 in nasal epithelium of AR patients and rats with AR. This may suggest that deficiency of MUC1 can be involved in the inflammation of the tonsils of allergic sensitized subjects. Further exploration of the genes more highly expressed in the AS subjects compared with NS subjects indicated that in addition to the changes in chemokines and MUC genes, IL-20RA gene is also upregulated. IL-20RA is an IL-10 related cytokine that is potentially associated to eczema. Higher expression of IL-20RA is associated with excessive cell proliferation and psoriasis such as skin disease.41 Since tonsils are made of several small crypts which are surrounded by epithelium, higher expression of IL-20RA indicates that it may play a role in sensitized tonsils.
The potentially most important and clinically relevant finding was the identification of upregulated genes in IL-17 pathway in AS versus NS and AS versus FS subjects. IL-17 is a proinflammatory cytokine well known for its immune protective function in several inflammatory diseases.42 This is also known to play crucial role in type-III inflammatory response against extracellular pathogens and fungi.43 A major function of IL-17 is the recruitment of neutrophils during an infection.44 Moreover, IL-17 induces the gene expression of some other genes in inflammatory response by activating immune pathways including NF-κB.44 However, the role of IL-17 pathway in human tonsils in relation to allergic sensitization has been unexplored previously. We identified that genes enriched in IL-17 pathways from our data are statistically significant; however, the gene ratio in this pathway is small but smaller alterations in the genes can alter the pathways.45 In addition, one gene can change and affect the gene expression, a group of genes expressed statistically significant, should not be ignored. NF-κB and MAPK signalling pathway are activated in NOD-like receptor (NLRs) signalling pathway. NLRs induce innate immunity by activating NF-κB and MAPK signalling pathways.46 Activation of these pathways induces the inflammatory cytokines or chemokines. NLRs are also considered intracellular sensors of microbial infection. In addition, NOD1 has been associated with asthma and atopic eczema.47 In our data, we identified the elevated expression of NOD-like receptor pathway that suggests the potential role of NLRs in tonsils of sensitized individuals.
The present study is the first one that has investigated the effect and association of variants with the allergic sensitization across the tonsillar transcriptome. The variant calling and subsequent expression quantitative trait locus (eQTL) analysis was performed to assess the presence of regulatory variants (cis or trans) that directly influence the expression of differentially expressed genes between atopic and non-atopic individuals.48 We observed 2 cis and 638 trans-eQTLs which have shown association with the expression of the DEGs in our data. Novel genes that were targets of these regulatory variants include the following: IL-1B, HLA-DQB1,49-51 MAPK3, MUC-21 and TUG1. Many of these genes are known candidates for allergic or tonsillar diseases revealed through genome-wide association studies (GWAS). For example, Bønnelykke et al52 has already described an association of HLA-DQB1 variant with allergic sensitization. Furthermore, present study also illustrates the complex regulation of IL-1B expression with largest number of trans-eQTLs (116, FDR <0.05). IL-1B is a member of cytokine family IL-1 which plays pivotal role in the activation of proinflammatory innate and adaptive immune response to a wide range of stimuli. IL-1B is very strong in exerting proinflammatory reactions to the extra cellular pathogens.53 Overall, these finding may suggest that these variants can have influence on the expression level of associated DEGs and with allergic sensitization.
The strengths of the study include carefully characterized patients and high laboratory standards. Our study has some limitations. All our study samples were taken from tonsillectomy patients suffering mainly from tonsillar hypertrophy or recurrent tonsillar infections. We did not have any samples from control/healthy subjects. Second, the ratio of the subject groups was slightly uneven, but the tools including DESeq2 enabled us to handle this problem quite well. DESeq2 distribute the read count to solve the batch problem before doing the differential expression analysis. Additionally, only subsets of patients had serum available for allergen specific IgE testing. Also, this study is entirely based on transcriptional profile and cannot identify potential cells involved. However, overall sample size of this study is supposed to decrease the bias. In addition, the findings of this study should be confirmed in an independent sample set.
5 CONCLUSIONS
The type of allergic sensitization is associated with extensive tonsillar transcriptomic alterations and changes in immune-related genes and pathways. Distinct differences were found between aeroallergen and food allergen sensitization groups, which could reflect different pathobiology behind them.
AUTHOR CONTRIBUTIONS
TJ and CA involved in conception and study design. TP, LI and EM collected the data. TH, FA, GT and LI involved in data management and analysis. TJ was a lead study coordinator. TH and TJ involved in drafting and writing of the manuscript. TJ, CA, STS, GT, FA, TH, LI, EM and TP involved in critical revision of the manuscript. All authors read, edited and approved the final manuscript.
ACKNOWLEDGMENTS
We thank Kirstin Jansen from SIAF for her contribution in making cytokine cluster in this study.
FUNDING INFORMATION
This work was supported by Sigrid Juselius foundation, Instrumenterium foundation, The Ella and Georg Ehrnrooth foundation, Finnish and Norweigen medical foundation, Maud Kuistila foundation, Ida Montin foundation, Vaino and Laina Kivi foundation, Biomedicum foundation, TYKS foundation, Päivikki and Sakari Sohlbergin foundation, Allergy Research Foundation and The Paulo foundation. L.I. was supported by the Finnish ORL-HNS Foundation and the Finnish Cultural Foundation. E.M. was supported by the Finnish Cultural Foundation. All in Finland.
CONFLICT OF INTEREST
The authors declare no conflict of interest in relation to this work.