Profiling of extracellular vesicle-bound miRNA to identify candidate biomarkers of chronic alcohol drinking in nonhuman primates
Sloan A. Lewis
Department of Molecular Biology and Biochemistry, University of California Irvine, Irvine, California, USA
Institute for Immunology, University of California Irvine, Irvine, California, USA
Search for more papers by this authorBrianna Doratt
Department of Molecular Biology and Biochemistry, University of California Irvine, Irvine, California, USA
Institute for Immunology, University of California Irvine, Irvine, California, USA
Search for more papers by this authorSuhas Sureshchandra
Department of Molecular Biology and Biochemistry, University of California Irvine, Irvine, California, USA
Institute for Immunology, University of California Irvine, Irvine, California, USA
Search for more papers by this authorTianyu Pan
Department of Statistics, University of California Irvine, Irvine, California, USA
Search for more papers by this authorSteven W. Gonzales
Oregon National Primate Research Center, Oregon Health and Science University, Beaverton, Oregon, USA
Search for more papers by this authorWeining Shen
Department of Statistics, University of California Irvine, Irvine, California, USA
Search for more papers by this authorKathleen A. Grant
Oregon National Primate Research Center, Oregon Health and Science University, Beaverton, Oregon, USA
Search for more papers by this authorCorresponding Author
Ilhem Messaoudi
Department of Molecular Biology and Biochemistry, University of California Irvine, Irvine, California, USA
Institute for Immunology, University of California Irvine, Irvine, California, USA
University of Kentucky, 760 Press Avenue, Lexington, United States, 40536-0679 USA
Correspondence
Ilhem Messaoudi, Microbiology, Immunology and Molecular Genetics, College of Medicine, University of Kentucky, 760 Press Avenue, Lexington, KY 40536
Email: [email protected]
Search for more papers by this authorSloan A. Lewis
Department of Molecular Biology and Biochemistry, University of California Irvine, Irvine, California, USA
Institute for Immunology, University of California Irvine, Irvine, California, USA
Search for more papers by this authorBrianna Doratt
Department of Molecular Biology and Biochemistry, University of California Irvine, Irvine, California, USA
Institute for Immunology, University of California Irvine, Irvine, California, USA
Search for more papers by this authorSuhas Sureshchandra
Department of Molecular Biology and Biochemistry, University of California Irvine, Irvine, California, USA
Institute for Immunology, University of California Irvine, Irvine, California, USA
Search for more papers by this authorTianyu Pan
Department of Statistics, University of California Irvine, Irvine, California, USA
Search for more papers by this authorSteven W. Gonzales
Oregon National Primate Research Center, Oregon Health and Science University, Beaverton, Oregon, USA
Search for more papers by this authorWeining Shen
Department of Statistics, University of California Irvine, Irvine, California, USA
Search for more papers by this authorKathleen A. Grant
Oregon National Primate Research Center, Oregon Health and Science University, Beaverton, Oregon, USA
Search for more papers by this authorCorresponding Author
Ilhem Messaoudi
Department of Molecular Biology and Biochemistry, University of California Irvine, Irvine, California, USA
Institute for Immunology, University of California Irvine, Irvine, California, USA
University of Kentucky, 760 Press Avenue, Lexington, United States, 40536-0679 USA
Correspondence
Ilhem Messaoudi, Microbiology, Immunology and Molecular Genetics, College of Medicine, University of Kentucky, 760 Press Avenue, Lexington, KY 40536
Email: [email protected]
Search for more papers by this authorFunding information
This study was supported by NIH 1R21AA025839-01A1 (Messaoudi), 5U01AA013510-20 (Grant), and 2R24AA019431-11 (Grant). S.A.L. is supported by NIH 1F31A028704-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Abstract
Background
Long-term alcohol drinking is associated with numerous health complications including susceptibility to infection, cancer, and organ damage. However, due to the complex nature of human drinking behavior, it has been challenging to identify reliable biomarkers of alcohol drinking behavior prior to signs of overt organ damage. Recently, extracellular vesicle-bound microRNAs (EV-miRNAs) have been found to be consistent biomarkers of conditions that include cancer and liver disease.
Methods
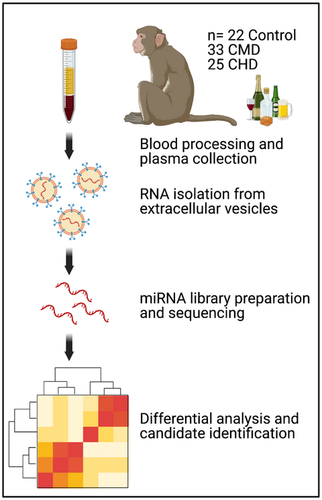
In this study, we profiled the plasma EV-miRNA content by miRNA-Seq from 80 nonhuman primates after 12 months of voluntary alcohol drinking.
Results
We identified a list of up- and downregulated EV-miRNA candidate biomarkers of heavy drinking and those positively correlated with ethanol dose. We overexpressed these candidate miRNAs in control primary peripheral immune cells to assess their potential functional mechanisms. We found that overexpression of miR-155, miR-154, miR-34c, miR-450a, and miR-204 led to increased production of the inflammatory cytokines TNFα or IL-6 in peripheral blood mononuclear cells after stimulation.
Conclusion
This exploratory study identified several EV-miRNAs that could serve as biomarkers of long-term alcohol drinking and provide a mechanism to explain alcohol-induced peripheral inflammation.
Graphical Abstract
Due to the complex nature of human drinking behavior, it has been challenging to identify reliable biomarkers of alcohol use that could be used to determine drinking behavior prior to signs of overt organ damage. This study profiles the plasma EV-miRNA content of 80 non-human primates after 12 months of voluntary ethanol drinking by miRNA-Seq. We identified several EV-miRNA that could serve as biomarkers of long-term alcohol drinking as well as provided a mechanism for alcohol-induced peripheral inflammation.
CONFLICT OF INTEREST
No competing interests reported.
Supporting Information
| Filename | Description |
|---|---|
| acer14760-sup-0001-FigS1.pdfPDF document, 162.9 KB | Fig S1 |
| acer14760-sup-0002-FigS2.pdfPDF document, 416.7 KB | Fig S2 |
| acer14760-sup-0003-FigS3.pdfPDF document, 452.3 KB | Fig S3 |
| acer14760-sup-0004-TableS1.xlsxExcel 2007 spreadsheet , 75.9 KB | Table S1 |
| acer14760-sup-0005-TableS2.xlsxExcel 2007 spreadsheet , 11.4 KB | Table S2 |
| acer14760-sup-0006-TableS3.xlsxExcel 2007 spreadsheet , 18.6 KB | Table S3 |
| acer14760-sup-0007-SupLegends.docxWord 2007 document , 14 KB | Supplementary Material |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
REFERENCES
- Alivernini, S., Gremese, E., McSharry, C., Tolusso, B., Ferraccioli, G., McInnes I.B. et al. (2018) MicroRNA-155—at the critical interface of innate and adaptive immunity in arthritis. Frontiers in Immunology, 8, http://doi.org/10.3389/fimmu.2017.01932
- Anttila, P., Järvi, K., Latvala, J. & Niemelä, O. (2004) Method-dependent characteristics of carbohydrate-deficient transferrin measurements in the follow-up of alcoholics. Alcohol and Alcoholism, 39, 59–63.
- Asquith, M., Pasala, S., Engelmann, F., Haberthur, K., Meyer, C., Park, B. et al. (2014) Chronic ethanol consumption modulates growth factor release, mucosal cytokine production, and microRNA expression in nonhuman primates. Alcoholism: Clinical and Experimental Research, 38, 980–993.
- Baker, E.J., Farro, J., Gonzales, S., Helms, C. & Grant, K.A. (2014) Chronic alcohol self-administration in monkeys shows long-term quantity/frequency categorical stability. Alcoholism: Clinical and Experimental Research, 38, 2835–2843.
- Bala, S., Csak, T., Kodys, K., Catalano, D., Ambade, A., Furi, I. et al. (2017) Alcohol-induced miR-155 and HDAC11 inhibit negative regulators of the TLR4 pathway and lead to increased LPS responsiveness of Kupffer cells in alcoholic liver disease. Journal of Leukocyte Biology, 102, 487–498.
- Barr, T., Girke, T., Sureshchandra, S., Nguyen, C., Grant, K. & Messaoudi, I. (2016) Alcohol consumption modulates host defense in rhesus macaques by altering gene expression in circulating leukocytes. The Journal of Immunology, 196, 182–195.
- Bartel, D.P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116, 281–297.
- Baum, M.K., Rafie, C., Lai, S., Sales, S., Page, J.B. & Campa, A. (2010) Alcohol use accelerates HIV disease progression. AIDS Research and Human Retroviruses, 26, 511–518.
- Bhattacharya, R. & Shuhart, M.C. (2003) Hepatitis C and alcohol: interactions, outcomes, and implications. Journal of Clinical Gastroenterology, 36, 242–252.
- Chang, L., Zhou, G., Soufan, O. & Xia, J. (2020) miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Research, 48, W244–W251.
- Conigrave, K.M., Degenhardt, L.J., Whitfield, J.B., Saunders, J.B., Helander, A., Tabakoff, B. et al. (2002) CDT, GGT, and AST as markers of alcohol use: the WHO/ISBRA collaborative project. Alcoholism: Clinical and Experimental Research, 26, 332–339.
- Djoussé, L., Gaziano, J.M. (2008) Alcohol consumption and heart failure: A systematic review. Current Atherosclerosis Reports, 10, (2), 117. –120. http://doi.org/10.1007/s11883-008-0017-z
- Fedirko, V., Tramacere, I., Bagnardi, V., Rota, M., Scotti, L., Islami, F. et al. (2011) Alcohol drinking and colorectal cancer risk: an overall and dose-response meta-analysis of published studies. Annals of Oncology, 22, 1958–1972.
- Ferracin, M., Lupini, L., Salamon, I., Saccenti, E., Zanzi, M.V., Rocchi, A. et al. (2015) Absolute quantification of cell-free microRNAs in cancer patients. Oncotarget, 6, 14545–14555.
- Grant, K.A., Leng, X., Green, H.L., Szeliga, K.T., Rogers, L.S. & Gonzales, S.W. (2008) Drinking typography established by scheduled induction predicts chronic heavy drinking in a monkey model of ethanol self-administration. Alcoholism: Clinical and Experimental Research, 32, 1824–1838.
- Grewal, P., Viswanathen, V.A. (2012) Liver cancer and alcohol. Clinics in Liver Disease, 16, (4), 839–850. http://doi.org/10.1016/j.cld.2012.08.011
- Groot, M. & Lee, H. (2020) Sorting mechanisms for microRNAs into extracellular vesicles and their associated diseases. Cells, 9, 1044.
- Hong, F., Kim, W.H., Tian, Z., Jaruga, B., Ishac, E., Shen, X. et al. (2002) Elevated interleukin-6 during ethanol consumption acts as a potential endogenous protective cytokine against ethanol-induced apoptosis in the liver: involvement of induction of Bcl-2 and Bcl-x(L) proteins. Oncogene, 21, 32–43.
- Huang, H.Y., Lin, Y.C., Li, J., Huang, K.Y., Shrestha, S., Hong, H.C. et al. (2020) miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Research, 48, D148–D154.
- Huang, X., Yuan, T., Tschannen, M., Sun, Z., Jacob, H., Du, M. et al. (2013) Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics, 14, 319.
- Hudolin, V. (1975) Tuberculosis and alcoholism. Annals of the New York Academy of Sciences, 252, 353–364.
- Jia, S., Zocco, D., Samuels, M.L., Chou, M.F., Chammas, R., Skog, J. et al. (2014) Emerging technologies in extracellular vesicle-based molecular diagnostics. Expert Review of Molecular Diagnostics, 14, 307–321.
- Jimenez, V.A., Helms, C.M., Cornea, A., Meshul, C.K. & Grant, K.A. (2015) An ultrastructural analysis of the effects of ethanol self-administration on the hypothalamic paraventricular nucleus in rhesus macaques. Frontiers in Cellular Neuroscience, 9, 260.
- Karatayli, E., Hall, R.A., Weber, S.N., Dooley, S. & Lammert, F. (2019) Effect of alcohol on the interleukin 6-mediated inflammatory response in a new mouse model of acute-on-chronic liver injury. Biochimica et Biophysica Acta – Molecular Basis of Disease, 1865, 298–307.
- Kline, S.E., Hedemark, L.L. & Davies, S.F. (1995) Outbreak of tuberculosis among regular patrons of a neighborhood bar. New England Journal of Medicine, 333, 222–227.
- Langmead, B. & Salzberg, S.L. (2012) Fast gapped-read alignment with Bowtie 2. Nature Methods, 9, 357–359.
- Lewis, S.A., Sureshchandra, S., Doratt, B., Jimenez, V.A., Stull, C., Grant, K.A. et al. (2021) Transcriptional, epigenetic, and functional reprogramming of monocytes from non-human primates following chronic alcohol drinking. Frontiers in Immunology, 12, 724015.
- Li, B., Liu, J., Xin, X., Zhang, L., Zhou, J., Xia, C. et al. (2021) MiR-34c promotes hepatic stellate cell activation and liver fibrogenesis by suppressing ACSL1 expression. International Journal of Medical Sciences, 18, 615–625.
- Lin, X., Rice, K.L., Buzzai, M., Hexner, E., Costa, F.F., Kilpivaara, O. et al. (2013) miR-433 is aberrantly expressed in myeloproliferative neoplasms and suppresses hematopoietic cell growth and differentiation. Leukemia, 27, 344–352.
- Malinski, M.K., Sesso, H.D., Lopez-Jimenez, F., Buring, J.E. & Gaziano, J.M. (2004) Alcohol consumption and cardiovascular disease mortality in hypertensive men. Archives of Internal Medicine, 164, 623–628.
- Messaoudi, I., Asquith, M., Engelmann, F., Park, B., Brown, M., Rau, A. et al. (2013) Moderate alcohol consumption enhances vaccine-induced responses in rhesus macaques. Vaccine, 32, 54–61.
- Miguez, M.J., Rosenberg, R., Burbano-Levy, X., Carmona, T. & Malow, R. (2012) The effect of alcohol use on IL-6 responses across different racial/ethnic groups. Future Virology, 7, 205–213.
- Momen-Heravi, F., Bala, S., Kodys, K. & Szabo, G. (2015) Exosomes derived from alcohol-treated hepatocytes horizontally transfer liver specific miRNA-122 and sensitize monocytes to LPS. Scientific Reports, 5, 9991.
- Mundle, G., Munkes, J., Ackermann, K. & Mann, K. (2000) Sex differences of carbohydrate-deficient transferrin, gamma-glutamyltransferase, and mean corpuscular volume in alcohol-dependent patients. Alcoholism: Clinical and Experimental Research, 24, 1400–1405.
- Natarajan, S.K., Pachunka, J.M. & Mott, J.L. (2015) Role of microRNAs in alcohol-induced multi-organ injury. Biomolecules, 5, 3309–3338.
- Odegaard, K.E., Chand, S., Wheeler, S., Tiwari, S., Flores, A., Hernandez, J. et al. (2020) Role of extracellular vesicles in substance abuse and HIV-related neurological pathologies. International Journal of Molecular Sciences, 21, 6765.
- Pfeifer, P., Werner, N. & Jansen, F. (2015) Role and function of microRNAs in extracellular vesicles in cardiovascular biology. BioMed Research International, 2015, 161393.
- Robinson, M.D., McCarthy, D.J. & Smyth, G.K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140.
- Saha, B., Momen-Heravi, F., Kodys, K. & Szabo, G. (2016) MicroRNA cargo of extracellular vesicles from alcohol-exposed monocytes signals naive monocytes to differentiate into M2 macrophages. Journal of Biological Chemistry, 291, 149–159.
- Saitz, R., Ghali, W.A. & Moskowitz, M.A. (1997) The impact of alcohol-related diagnoses on pneumonia outcomes. Archives of Internal Medicine, 157, 1446–1452.
- Sesso, H.D., Cook, N.R., Buring, J.E., Manson, J.E., Gaziano, J.M. (2008) Alcohol Consumption and the Risk of Hypertension in Women and Men. Hypertension, 51, (4), 1080. –1087. http://doi.org/10.1161/hypertensionaha.107.104968
- Sureshchandra, S., Rais, M., Stull, C., Grant, K. & Messaoudi, I. (2016) Transcriptome profiling reveals disruption of innate immunity in chronic heavy ethanol consuming female rhesus macaques. PLoS One, 11, e0159295.
- Sureshchandra, S., Raus, A., Jankeel, A., Ligh, B.J.K., Walter, N.A.R., Newman, N. et al. (2019) Dose-dependent effects of chronic alcohol drinking on peripheral immune responses. Scientific Reports, 9, 7847.
- Szabo, G. & Bala, S. (2010) Alcoholic liver disease and the gut-liver axis. World Journal of Gastroenterology, 16, 1321–1329.
- Szabo, G. & Saha, B. (2015) Alcohol's effect on host defense. Alcohol Research, 37, 159–170.
- Torres, J.L., Novo-Veleiro, I., Manzanedo, L., Alvela-Suárez, L., Macías, R., Laso, F.J. et al. (2018) Role of microRNAs in alcohol-induced liver disorders and non-alcoholic fatty liver disease. World Journal of Gastroenterology, 24, 4104–4118.
- Valadi, H., Ekström, K., Bossios, A., Sjöstrand, M., Lee, J.J, Lötvall, J.O. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology, 9, (6), 654. –659. http://doi.org/10.1038/ncb1596
- Viel, G., Boscolo-Berto, R., Cecchetto, G., Fais, P., Nalesso, A. & Ferrara, S.D. (2012) Phosphatidylethanol in blood as a marker of chronic alcohol use: a systematic review and meta-analysis. International Journal of Molecular Sciences, 13, 14788–14812.
- Wu, J., Li, X., Li, D., Ren, X., Li, Y., Herter, E.K. et al. (2020) MicroRNA-34 family enhances wound inflammation by targeting LGR4. The Journal of Investigative Dermatology, 140, 465–476.e11.
- Yang, Z., Tsuchiya, H., Zhang, Y., Hartnett, M.E. & Wang, L. (2013) MicroRNA-433 inhibits liver cancer cell migration by repressing the protein expression and function of cAMP response element-binding protein. Journal of Biological Chemistry, 288, 28893–28899.
- Zhang, Y., Jia, Y., Zheng, R., Guo, Y., Wang, Y., Guo, H. et al. (2010) Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clinical Chemistry, 56, 1830–1838.
- Zhou, Y., Zhou, B., Pache, L., Chang, M., Khodabakhshi, A.H., Tanaseichuk, O. et al. (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature Communications, 10, 1523.