Comprehensive genetic analysis of histological components of combined small cell carcinoma
Funding information: JSPS KAKENHI, Grant/Award Number: 19K17651
Abstract
Background
Combined small-cell lung cancer (cSCLC) is a rare type of small-cell lung cancer (SCLC) that includes both SCLC and non-small-cell lung cancer (NSCLC). The molecular biological mechanisms underlying the heterogeneity of histological types in combined or metachronously transformed SCLC (mtSCLC) remain unclear. This study aimed to investigate the relationship between genetic alterations and each histological component heterogeneously detected in cSCLC and mtSCLC.
Methods
This study included four cSCLC cases and one mtSCLC case. Formalin-fixed and paraffin-embedded sections of each histological component of these tumors were subjected to next-generation sequencing (NGS) and quantitative reverse transcription-polymerase chain reaction to investigate the genetic mutations and expression levels of neuroendocrine cell-specific transcription factors (achaete-scute homolog-1 [ASCL1], brain-2 [BRN2] also known as POU domain class 3 transcription factor 2, nuclear factor 1 B [NF1B], insulinoma-associated protein 1 [INSM1], and thyroid transcription factor-1 [TTF-1]).
Results
NGS analysis revealed that SCLC and NSCLC components share the same somatic mutations detected most frequently in TP53, and also in RB1 and EGFR. Gene expression analysis showed ASCL1 expression was significantly lower in the NSCLC component than in the SCLC component.
Conclusion
We conclude that the morphological evolution of heterogeneous histological components in cSCLC may be associated with differences in ASCL1 expression levels, but not in acquired somatic gene mutations.
INTRODUCTION
Small-cell lung cancer (SCLC) is a highly aggressive neuroendocrine tumor that accounts for 13% of all lung cancers worldwide.1 Combined SCLC (cSCLC) is a type of SCLC with an incidence of 28% of all SCLC cases diagnosed by surgical specimens.2 The World Health Organization (WHO) Tumor Classification defines it as SCLC combined with additional components of any histological type of non-small-cell lung carcinoma (NSCLC).3 Cases of NSCLC have been described with epidermal growth factor receptor (EGFR) gene mutation that was metachronously transformed to SCLC after treatment with EGFR-tyrosine kinase inhibitors (TKIs).4, 5 However, the cell origin of SCLC and NSCLC is usually thought to be different: SCLC generally arises from neuroendocrine cells or neuroendocrine progenitors, whereas adenocarcinoma originates from alveolar type 2 and club cells.6, 7 Another report indicated that alveolar type II cells might form SCLC, although less frequently than neuroendocrine cells.8 The cell origin of cSCLC and metachronously transformed SCLC (mtSCLC) underlying the heterogeneity of different histological components is unknown.
The genetic background of the two histological components of cSCLC is unclear. Only a few reports have investigated the genetic profiles of different histologic components in cSCLC using next-generation sequencing (NGS).9, 10
Achaete-scute homolog-1 (ASCL1), a basic helix–loop–helix transcription factor, is necessary to initiate the development of SCLC in a mouse model and induce neuroendocrine differentiation in SCLC.11-13 The transcription factor nuclear factor 1 B (NF1B) is targeted by ASCL1 and drives tumor initiation and progression in mouse models of SCLC.14, 15 Insulinoma-associated protein 1 (INSM1) is a zinc-finger transcription factor. It is a crucial regulator of ASCL1, brain-2 (BRN2), and neuroendocrine molecules in lung cancer cells and plays a role in the proliferation and apoptosis of SCLC.16 Thyroid transcription factor-1 (TTF-1) expression levels have been associated with neuroendocrine differentiation via the expression of its regulators, such as ASCL1 and NF1B, in SCLC.17 The roles of these transcription factors in the histological differences in cSCLC and mtSCLC have not been elucidated. This study aimed to clarify the status of genetic mutations and gene expressions related to morphological heterogeneity in cSCLC and mtSCLC.
METHODS
Patients
Patients with cSCLC, mtSCLC, and pure SCLC consecutively diagnosed and treated at the Nihon University Itabashi Hospital (Tokyo, Japan) between 2010 and 2019 were enrolled in this study. The study design was approved by the Institutional Review Board (265-0, 30-14-0) according to the Declaration of Helsinki. The study investigated somatic mutations in targeted cancer panels; germline mutations were excluded.
Patients' clinical information was extracted from the medical records at Nihon University Itabashi Hospital (Table 1). Diagnoses of cSCLC, mtSCLC, and pure SCLC were based on the 2021 WHO classification of lung tumors3 by trained histopathologists. Four patients (cases 1–4) had limited cSCLC. One patient (case 5) had advanced mtSCLC that had progressed from adenocarcinoma. The FFPE tissues of primary and/or metastatic lesions of these patients were subjected to immunohistochemical analysis. Three cSCLC samples (cases 1–3) and one mtSCLC sample (case 5) were subjected to NGS analysis, and four cSCLC samples (cases 1–4) and six pure SCLC samples (Table S1) were subjected to quantitative RT-PCR analyses.
| Cases | Synchronous/metachronous | Histological component | Stage | Sex | Age | Smoking | Treatment | Follow-up |
|---|---|---|---|---|---|---|---|---|
| 1 | Synchronous | SC/Ad | IA | F | 76 | + | Surgery | Died 20 months after diagnosis |
| Adjuvant chemotherapy | ||||||||
| 2 | Synchronous | SC/Sq | IIB | M | 81 | + | Surgery | Transferred 6 months after diagnosis |
| 3 | Synchronous | SC/Ad | IB | M | 60 | + | Surgery | Alive 38 months after diagnosis |
| Adjuvant chemotherapy | ||||||||
| 4 | Synchronous | SC/Sq | IIIB | M | 76 | + | surgery | No information |
| Adjuvant chemotherapy | ||||||||
| 5 | Metachronous | SC/Ad | IV | F | 70 | − | EGFR-TKI (before SC) | Transferred 73 months after diagnosis |
| Chemotherapy (after SC) |
- Abbreviations: Ad, adenocarcinoma; EGFR-TKI, epidermal growth factor receptor-tyrosine kinase inhibitor; SC, small-cell lung cancer; Sq, squamous cell carcinoma.
Immunohistochemistry
The expressions of CD56, synaptophysin, chromogranin A, ASCL1, INSM1, TTF-1, and Ki-67 were evaluated using immunohistochemistry. The 4-μm thick sections were mounted on silane-coated glass slides. After deparaffinization, samples were boiled in citrate buffer (pH 6.0) for CD56, synaptophysin, and chromogranin A, and in ethylenediamine tetraacetic acid buffer (pH 9.0) for ASCL1, INSM1, and TTF-1, as antigen retrieval. Subsequently, an automated staining system (Histostainer; Nichirei Bioscience, Tokyo, Japan) was used for immunostaining, which was performed as follows: (1) blocking with 3% hydrogen peroxide to eliminate endogenous peroxidase for 5 min at room temperature; (2) incubation with 1/50 primary mouse monoclonal anti-Ki-67 antibody (clone MIB-1; Agilent Technologies Inc.), 1/50 mouse monoclonal anti-CD56 antibody (clone 123-C3, Agilent Technologies Inc.), 1/100 mouse monoclonal anti-chromogranin A antibody (clone DAK-A3, Agilent Technologies Inc.), 1/50 rabbit polyclonal anti-synaptophysin antibody (Agilent Technologies Inc.), 1/1000 rabbit monoclonal anti-ASCL1 antibody (clone EPR19592; Abcam PLC), 1/100 mouse monoclonal anti-INSM1 antibody (clone A-8; Santa Cruz Biotechnology Inc.), and 1/100 mouse monoclonal anti-TTF-1 antibody (clone 8G7G3/1; Agilent Technologies) each for 30 min at room temperature; (3) washing with phosphate buffered saline (PBS); (4) incubation with the polymer second antibody (Simple stain Max PO Multi; Nichirei Bioscience) for 30 min at room temperature; (5) washing with PBS; (6) dyeing with 3,3-diaminobenzidine for 10 min at room temperature; (7) washing with PBS; (8) counterstaining with hematoxylin; and (9) dehydrating and covering with cover glasses. The detailed methods have been previously reported.17
Total DNA extraction, targeted enrichment, and sequencing
The 8-μm thick sections were mounted on regular glass slides. After deparaffinization, the target tumor cells were macro-dissected and collected in a 1.5-mL tube. The target tumor cells were identified using hematoxylin–eosin (HE) staining. Genomic DNA was extracted using the QIAamp DNA FFPE Tissue Kit (Qiagen) according to the manufacturer's instructions. The concentration and quality of the extracted DNA were measured using the GeneRead DNA QuantiMIZE Kit (Qiagen) before the targeted enrichment. The coding regions and exon/intron junctions of 72 oncogenes were enriched by multiplex PCR using the QIAseq Targeted DNA Human Lung Cancer Panel (DHS-005Z; Qiagen) according to the manufacturer's instructions and sequenced using NextSeq 500 (Illumina) in 151-base pair (bp) paired-end reads. The average read depth of coverage was set at 3400× to allow the detection of rare mutations and to accurately estimate variant allele frequencies.
Alignment and somatic variant calling
Alignment and somatic variant calling were performed using the Qiagen Data Analysis Center web service (https://ngsdataanalysis.qiagen.com/QIAseqDNA). Original FASTQ files generated by NextSeq were uploaded to the web service. Subsequently, smCounter v2 was used for somatic variant calling.18
Variant annotation and filtering
Functional annotations of the Ensembl database GRCh37.7519 and the possible effects of variants were added using SnpEff version 4.2.20 Using these annotations, variants were filtered first for those that were predicted to alter amino acid sequences (missense, nonsense, and splice-site mutations and indels in coding regions), then for those that were rare (<1.0% minor allele frequencies in the HapMap-JPT [Japanese in Tokyo, Japan], the 1000 Genomes EAS [East Asian population including 104 Japanese individuals], or the Human Genetic Variation Database [http://www.genome.med.kyoto-u.ac.jp/SnpDB/]). Furthermore, we used the genome data of eight previously sequenced healthy Japanese men aged over 70 years with no relevant medical history for variant filtering. Those with a variant allele frequency (VAF) ≥10% were targeted. These mutations were validated and analyzed in normal lung tissues using Sanger sequencing.
Total RNA extraction and cDNA synthesis
The 8-μm thick sections were mounted on regular glass slides. After deparaffinization, the target tumor cells, identified using HE staining, were dissected and collected in 1.5-mL tubes. Total RNA was extracted using an RNeasy FFPE Kit (Qiagen) according to the manufacturer's instructions. The RNA samples were dissolved in 5 μl of RNase-free water, and the concentration was measured using the Nanodrop (Thermo Fisher Scientific Inc.). Total RNA samples were stored at −80°C until use. Genomic DNA was eliminated and cDNA was synthesized using the QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer's instructions.
Quantitative RT-PCR assay
We investigated the mRNA expression levels of TTF-1, ASCL1, BRN2, NF1B, INSM1, and the internal control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), using quantitative RT-PCR. Quantitative RT-PCR was performed using 1 μl of cDNA, TaqMan probes (Thermo Fisher Scientific Inc.), and a Step One Plus quantitative RT-PCR machine (Thermo Fisher Scientific Inc.). The premixed primers and probes were as follows: ASCL1 (Assay ID: Hs04187546_g1), BRN2 (Assay ID: Hs00271595_s1), NF1B (Assay ID: Hs01029175_m1), TTF-1 (Assay ID: Hs00968940_m1), INSM1 (Assay ID: Hs00357871_s1), and GAPDH (Assay ID: Hs99999905_m1). The quantitative RT-PCR thermal cycling profile was as follows: 95°C for 10 min, followed by 50 cycles of 95°C for 15 s and 60°C for 1 min. The expression levels of the target mRNA were calculated using the ΔCt method, with GAPDH mRNA expression as the reference.21
Statistical analysis
The correlations between the components of SCLC and NSCLC in cSCLC and mtSCLC observed by immunohistochemistry (with respect to the markers synaptophysin, chromogranin A, CD56, ASCL1, INSM1, and TTF-1) were analyzed using Fisher's exact test. Correlations in terms of the Ki-67 expression level between components of SCLC and NSCLC in cSCLC and mtSCLC were analyzed using the Mann–Whitney U test. The differences in mRNA expression levels among pure SCLC, SCLC component, and NSCLC component in cSCLC and mtSCLC were analyzed using the Kruskal–Wallis test. The Steel–Dwass post-hoc test was used for multiple comparisons. All statistical analyses were performed using EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan), a graphical interface for R (The R Foundation for Statistical Computing).22
RESULTS
Immunohistochemical findings
Immunohistochemical neuroendocrine and proliferation markers were evaluated in the SCLC and NSCLC components (Figure 1). The SCLC component showed higher positive rates for synaptophysin, CD56, ASCL1, and INSM1, and a higher labeling index for Ki67 compared to the NSCLC component (Table 2 and Figure S1). The significant difference in the Ki-67 labeling index (%) was higher in the SCLC component (mean 72%) than in the NSCLC component (mean 30%). There were no significant differences in chromogranin A or TTF-1 expression (Table 2).
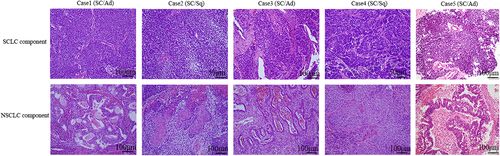
| Case | Histological component | Syn | Chromo | CD56 | ASCL1 | TTF-1 | INSM1 | Ki 67 labeling index (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | SC | + | − | + | − | + | − | 90 |
| Ad | − | − | − | − | + | − | 20 | |
| 2 | SC | + | − | + | ± | + | ± | 80 |
| Sq | − | − | − | − | − | − | 30 | |
| 3 | SC | + | + | + | + | ++ | + | 80 |
| Ad | − | − | − | − | ++ | − | 70 | |
| 4 | SC | ++ | ++ | ++ | ++ | − | ++ | 30 |
| Sq | − | − | ± | − | − | − | 10 | |
| 5 | SC | ++ | − | ++ | + | ++ | ++ | 80 |
| Ad | − | − | ++ | ± | ++ | − | 70 | |
| Total | SC | 5/5** | 2/5 | 5/5* | 4/5* | 4/5 | 4/5* | 72 (30–90)* |
| NSC | 0/5 | 0/5 | 2/5 | 1/5 | 3/5 | 0/5 | 30 (10–70) |
- * p < 0.05.
- ** p < 0.01.
- Total Ki67 labeling index is shown as mean and range.
- Abbreviations: Ad, adenocarcinoma; Chromo, chromogranin A; NSC, non-small-cell carcinoma; SC, small-cell carcinoma; Sq, squamous cell carcinoma; Syn, synaptophysin.
- Immunohistochemical evaluations: −, negative; ±, rare; +, focal; ++, diffuse.
- The Total column shows the positivity/total cases for each immunohistochemical antibody between small-cell carcinoma and non-small-cell carcinoma.
Genetic mutations in SCLC and NSCLC components
Genetic mutations in three cSCLC cases and one mtSCLC case were analyzed using samples with sufficient levels of DNA quality and volume. The tumor contents comprised 80–90% of the SCLC component and 20–80% of the NSCLC component. Table 3 lists the somatic mutations that were detected in each case. Figure S2 shows the somatic mutations validated using Sanger sequencing. In four cases, 17 mutations in nine genes (EGFR, RB1, TP53, MUC16, SMARCA4, KDR, PKHD1, KMT2D, and RBM10) were found. The most common mutation type was the missense mutation (88%, 15/17 mutations). According to the ClinVar database, two pathogenic somatic mutations were clinically significant: p.Ser768Ile in EGFR (case 1) and p.Arg251* in RB1 (case 5), one pathogenic/likely pathogenic somatic mutation, p.Arg158His in TP53 (case 1), and one likely pathogenic somatic mutation, p.Arg175Leu in TP53 (case 5). A drug response variant was also detected (p.Leu858Arg in EGFR [case 5]). The somatic mutations with uncertain significance were p.Pro142Leu in TP53 (case 5), p.Ala159Val in TP53 (case 2), p.Lys1540Arg in SMARCA4 (case 5), p.Val774Met in EGFR (case 1), and p.Glu654Lys in PKHD1 (case 2).
| Cases | Tumor content rate | Syn/Meta | Type | VAF | Gene | Mutation type | HGVSc | HGVSp | ClinVar interpretation | Sample no.a |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | SC: 90% Ad: 20% |
Syn | SC | 0.6505 | EGFR | Missense | c.2303G > T | p.Ser768Ile | Pathogenic | 1 |
| Ad | 0.4727 | 2 | ||||||||
| SC | 0.6504 | EGFR | Missense | c.2320G > A | p.Val774Met | Uncertain significance | 3 | |||
| Ad | 0.4741 | 4 | ||||||||
| SC | 0.7796 | TP53 | Missense | c.473G > A | p.Arg158His | Pathogenic/Likely pathogenic | 5 | |||
| Ad | 0.5872 | 6 | ||||||||
| 2 | SC:90% Sq: 80% |
Syn | SC | 0.6372 | KDR | Missense | c.1292C > T | p.Pro431Leu | 7 | |
| Sq | 0.6513 | 8 | ||||||||
| SC | 0.6406 | PKHD1 | Missense | c.1960G > A | p.Glu654Lys | Uncertain significance | 9 | |||
| Sq | 0.6868 | 10 | ||||||||
| SC | 0.6107 | PKHD1 | Missense | c.1139 T > C | p.Phe380Ser | 11 | ||||
| Sq | 0.6531 | 12 | ||||||||
| SC | 0.4899 | KMT2D | Splice donor/intron | c.16052 + 1G > A | 13 | |||||
| Sq | 0.5891 | 14 | ||||||||
| SC | 0.8426 | TP53 | Missense | c.476C > T | p.Ala159Val | Uncertain significance | 15 | |||
| Sq | 0.8609 | 16 | ||||||||
| 3 | SC: 80% Ad: 50% |
Syn | SC | 0.6042 | RB1 | Missense/splice region | c.861G > T | p.Glu287Asp | 17 | |
| SC | 0.5063 | MUC16 | missense | c.4539 T > G | p.Asp1513Glu | 18 | ||||
| Ad | 0.4321 | 19 | ||||||||
| SC | 0.7765 | RBM10 | Missense | c.920 T > A | p.Leu307Gln | 20 | ||||
| 5 | SC: 80% Ad: 70% |
Meta | SC | 0.8944 | EGFR | Missense | c.2573 T > G | p.Leu858Arg | Drug response | 21 |
| Ad | 0.6461 | 22 | ||||||||
| SC | 0.9450 | RB1 | Nonsense | c.751C > T | p.Arg251* | Pathogenic | 23 | |||
| Ad | 0.7846 | 24 | ||||||||
| SC | 0.9353 | TP53 | Missense | c.524G > T | p.Arg175Leu | Likely pathogenic | 25 | |||
| Ad | 0.6989 | 26 | ||||||||
| SC | 0.9595 | TP53 | Missense | c.425C > T | p.Pro142Leu | Uncertain significance | 27 | |||
| Ad | 0.7290 | 28 | ||||||||
| Ad | 0.3437 | MUC16 | Missense | c.37181 T > A | p.Phe12394Tyr | 29 | ||||
| SC | 0.6614 | SMARCA4 | Missense | c.4619A > G | p.Lys1540Arg | Uncertain significance | 30 | |||
| Ad | 0.6537 | 31 |
- Abbreviations: Ad, adenocarcinoma; HGVS, Human Genome Variation Society; Meta, metachronous; SC, small-cell lung cancer; Sq, squamous cell carcinoma; Syn, synchronous.
- a Sample no. corresponds to Figure S1.
In each case, most genetic mutations were shared between SCLC and NSCLC components: 3/3 (100%) in case 1, 5/5 (100%) in case 2, 1/3 (33%) in case 3, and 5/6 (83%) in case 5. The common shared genetic mutations were TP53 (3/4 cases), EGFR (2/4 cases), KDR, PKHD1, KMT2D, MUC16, RB1, and SMARCA4 (1/4 cases).
mRNA expression levels of ASCL1, BRN2, NF1B, TTF-1, and INSM1 in the SCLC and NSCLC components of cSCLC and pure SCLC
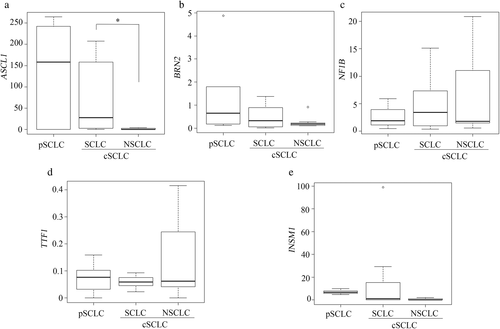
The mRNA expression levels of ASCL1 were significantly lower in the NSCLC component than in the SCLC component of cSCLCs (p = 0.029; Figure 2a). The ASCL1 expression level tended to be higher in the pure SCLC than in SCLC components of cSCLC, although this was not statistically significant (Figure 2a). BRN2, NF1B, TTF-1, and INSM1 expression levels did not differ significantly between the pure SCLC and SCLC components or between the SCLC and NSCLC components of cSCLC (Figure 2b–e).
DISCUSSION
The present study demonstrated that cSCLC and mtSCLC share the same major gene mutations in the SCLC and NSCLC components. However, in the NSCLC component, the ASCL1 expression was significantly lower than in the SCLC component of the cSCLC.
Similar to our result, a previous study also showed that approximately 75% of the identified somatic mutations, such as TP53, were present in both components of three cSCLC cases.9 These results suggest that SCLC and NSCLC components in cSCLC originate from a common ancestor because they have a similar genetic background. In this study, the most common gene mutation was the TP53 mutation found in all three cases. The TP53 mutations have been reported in approximately 90% of SCLC cases and 46% of NSCLC (adenocarcinoma) cases.23, 24 Furthermore, shared RB1 mutations were observed in one case. RB1 mutations have been reported in approximately 65% of SCLC cases and 4% of NSCLC (adenocarcinoma) cases.23, 24 Bi-allele TP53 and RB1 mutations are early and necessary key events in the development of pure SCLC in humans.14, 25 The same TP53 and RB1 mutations were found in both SCLC and NSCLC histological components in this study. From the results of these gene mutations, the cell origin of each histological component of cSCLC may be closer to that of pure SCLC than that of NSCLC. In addition, pathogenic EGFR mutations were shared by both SCLC and NSCLC. Previous reports have shown that different histological components harbor the same EGFR mutation in transformation into SCLC as a mechanism of resistance to EGFR TKIs.5, 26 In this study, one cSCLC case had an EGFR mutation in both histological components, independent of transformation into SCLC as a mechanism of resistance to EGFR TKIs. Since there have been previous reports that cSCLC responds to EGFR-TKI, it may be important to perform an EGFR gene mutation analysis in cSCLC to obtain additional therapeutic options.26
ASCL1 regulates neuroendocrine differentiation; in particular, it upregulates synaptophysin and contributes to proliferation and migration by targeting cyclin-dependent kinase 5 in SCLC.27-30 In the present study, the more frequent immunohistochemical positivity for synaptophysin, CD56, ASCL1, and INSM1 and a higher labeling index for Ki67 in the SCLC were correlated with higher ASCL1 expression. Our results support the previous reports and suggest that ASCL1 plays an important role in the development of SCLC. Interestingly, ASCL1 expression tended to be highest in pure SCLC and was lower in the NSCLC component than in the SCLC component. Cases that did not express ASCL1 and NEUROD1 in cSCLC compared to pure SCLC have been described.31 Decreasing ASCL1 expression in the NSCLC component is considered to be associated with the activation of NOTCH signaling regulated by histone modification and differentiation to NSCLC from SCLC.32 Another report hypothesized that cSCLC might originate from pure SCLC, partially decreasing ASCL1 in the NSCLC component.33 Our results suggest that morphological transformation into NSCLC from SCLC might occur depending on decreased ASCL1 expression, but not because of genetic mutations, because of the presence of highly similar genetic backgrounds between the SCLC and NSCLC components (Figure 3).
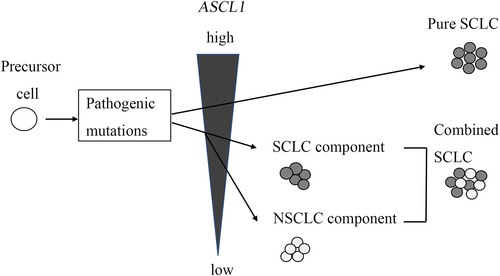
In the present series, there was a peculiar case with lower ASCL1 expression in both SCLC and NSCLC components (case 1). SCLC subtypes are recently defined by differential expression of four key transcription regulators: ASCL1, NEUROD1, yes-associated protein 1 (YAP1) and POU2F3.34 SCLCs with low expression of neuroendocrine markers are classified as YAP1 or POU2F3 types. Whether the cSCLC of case1 represents the YAP1 or POU2F3 type remains to be addressed. SCLC subtyping is more important clinically because the therapeutic potential of inhibitors targeting delta-like protein 3 (DLL3), an actionable target of ASCL1, is currently under further investigation in clinical trials.35 In ASCL1-high SCLCs, DLL3-targeting therapy is predicted to be more effective owing to the higher expression of DLL3.36 Our results suggest that targeted therapy may be less effective in cSCLC than in pure SCLC because of lower levels of ASCL1. Further clinical investigations of cSCLC are necessary.
This study has some limitations. First, the sample size was small. The small sample size might make it difficult to clarify the expression levels of NF1B, BRN2, TTF-1, and INSM1. Second, the sensitivity of the methodologies may have influenced the results. The genetic mutations found in NGS were validated using Sanger sequencing, which has lower sensitivity than NGS, therefore gene mutations that could not be verified by Sanger sequencing could not be identified. Third, germline mutations were not analyzed in the present study. One mutation (case 3, MUC16) was found in cancerous and noncancerous lung tissues as a secondary finding, which might have resulted from a mixture of cancerous tissues or a germline mutation; however, the ClinVar database has no information regarding this mutation. Thus, this mutation warrants further investigation. Finally, a small number of different histological components might have been admixed. However, each component was collected separately as much as possible via macrodissection.
In conclusion, this study demonstrated that each histological component in SCLC may have morphological evolution depending on the difference in ASCL1 expression, not due to the differences in acquired somatic mutations.
ACKNOWLEDGMENTS
We would like to thank DNA Chip Research Inc. for their technical assistance.
DISCLOSURE
The authors have no conflicts of interest to declare.
FUNDING
This work was supported by JSPS KAKENHI (Grant Number: 19 K17651).