Genomic analysis between idiopathic pulmonary fibrosis and associated lung cancer using laser-assisted microdissection: A case report
Abstract
Lung cancer (LC) is the most fatal complication of idiopathic pulmonary fibrosis (IPF). However, the molecular pathogenesis of the development of LC from IPF is still unclear. Here, we report a case of IPF-associated LC for which we investigated the genetic alterations between IPF and LC. We extracted formalin-fixed paraffin-embedded DNA from each part of the surgical lung tissue using a laser-assisted microdissection technique. The mutations in each part were detected by next-generation sequencing (NGS) using 72 lung cancer-related mutation panels. Five mutations were found in IPF and four in LC. Almost all somatic mutations did not overlap between the IPF and LC regions. These findings suggest that IPF-associated LC may not be a result of the accumulation of somatic mutations in the regenerated epithelium of the honeycomb lung in the IPF region.
INTRODUCTION
Idiopathic pulmonary fibrosis (IPF) is a chronic, progressive fibrosing interstitial pneumonia that occurs primarily in older adults. It has a short median survival of three years following diagnosis.1, 2 Lung cancer (LC) is one of the most fatal comorbidities of IPF, with a prevalence of 4.4 to 48% in patients with IPF.2 Previous reports have shown that point somatic mutations in the K-ras and TP53 genes in the lung tissue of IPF-associated LC may arise from regenerated alveolar epithelial cells and can lead to the development of LC.3 Some IPF-associated LC is associated with potentially targetable alterations such as BRAF mutations.4 However, the detailed genomic mechanism of the pathogenesis of IPF-associated LC remains unclear, and a specific targeted treatment has not been developed.
In this report, we investigated the differences in genetic mutations between the honeycomb lung and coexisting LC in an IPF-associated LC patient with next-generation sequencing (NGS) analysis using laser-assisted microdissection (LMD).
CASE REPORT
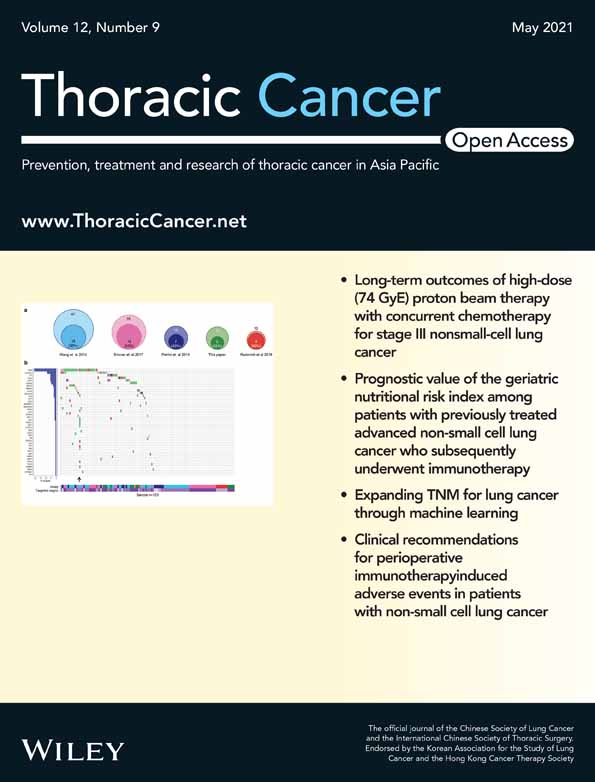
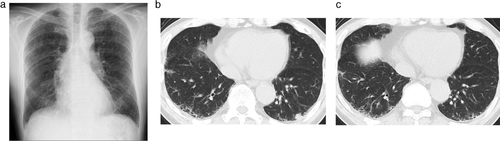
A 69-year-old Japanese man was diagnosed with squamous cell carcinoma T1bN0M0 in the left lower lung one year after being diagnosed with IPF (Figure 1). Partial resection of the left lower lobe was performed. For clinical research purposes, the tumor DNA was extracted from the LC and epithelial cells of honeycomb lung formalin-fixed paraffin-embedded (FFPE) tissue (Figure 2) In particular, the epithelial cells in the honeycomb lung in IPF were collected using LMD (Figure 2). Sequencing analysis was performed with targeted NGS using an LC-related mutation panel (Table 1). Using NGS analysis, five mutations (KDR, EPHA5, APC, CREBBP, and ERBB2) were detected in the IPF sample, and four mutations (EPHA5, PKHD1, RB1, and KEAP1) were detected in the LC sample (Table 2). Although one mutation (EPHA5) overlapped between the IPF and LC samples, there was no overlap between the other mutations.
| AKT1 | CDKN2B | FGFR1 | KMT2D | NF1 | PIK3R1 | RIT1 | TSC1 |
|---|---|---|---|---|---|---|---|
| ALK | CREBBP | FGFR2 | KRAS | NFE2L2 | PIK3R2 | ROS1 | U2AF1 |
| AMER1 | CTNNB1 | FGFR3 | LRP1B | NOTCH1 | PKHD1 | RUNX1T1 | |
| APC | DDR2 | FHIT | MAP2K1 | NRAS | PTEN | SETD2 | |
| ARID1A | EGFR | GRM8 | MDM2 | NTRK1 | PTPRD | SMAD4 | |
| ATM | EPHA5 | HRAS | MET | NTRK2 | RARB | SMARCA4 | |
| BAI3 | ERBB2 | JAK2 | MGA | NTRK3 | RASSF1 | SOX2 | |
| BAP1 | ERBB4 | KDR | MLH1 | PDGFRA | RB1 | STK11 | |
| BRAF | FBXO7 | KEAP1 | MUC16 | PIK3CA | RBM10 | TNFAIP3 | |
| CDKN2A | FBXW7 | KIT | MYC | PIK3CG | RET | TP53 | |
| No. | Chr | Position | Mutation type | Gene | HGVS.c | HGVS.p | Clinical significance | VAF | ||
|---|---|---|---|---|---|---|---|---|---|---|
| IPF | LC | |||||||||
| 1 | IPF | 4 | 55 955 965 | missense_variant | KDR | c.3197G>A | p.Arg1066His | No information | 0.021 | |
| 2 | IPF, LC | 4 | 66 231 683 | missense_variant | EPHA5 | c.2017T>A | p.Ser673Thr | Probably benign | 0.459 | 0.596 |
| 3 | IPF | 5 | 112 162 877 | missense_variant | APC | c.1481G>T | p.Ser494Ile | No information | 0.034 | |
| 4 | LC | 6 | 51 612 932 | missense_variant | PKHD1 | c.9482T>C | p.Val3161Ala | No information | 0.256 | |
| 5 | LC | 13 | 48 954 198 | stop_gained | RB1 | c.1399C>T | p.Arg467* | Pathogenic | 0.339 | |
| 6 | IPF | 16 | 3 860 664 | missense_variant | CREBBP | c.915C>G | p.Asn305Lys | No information | 0.032 | |
| 7 | IPF | 17 | 37 873 598 | missense_variant | ERBB2 | c.1763C>T | p.Ala588Val | No information | 0.022 | |
| 8 | LC | 19 | 10 602 620 | missense_variant | KEAP1 | c.958C>T | p.Arg320Trp | No information | 0.380 | |
- Note: Content rate of the target cells; IPF:100.0%, LC:78.7%. *: Stop codon.
- Abbreviations: Chr, chromosome; HGVS, human genome variation society; IPF, idiopathic pulmonary fibrosis; LC, lung cancer; VAF, variant allele frequency.
DISCUSSION
IPF is a chronic, progressive, and fatal fibrosing interstitial pneumonia of unknown cause.1 LC is a major comorbidity of IPF and has a significant adverse impact on the survival of IPF patients.5
IPF and LC share common pathogenetic features, such as increased proliferation rates, immune dysregulation, resistance to apoptosis, telomere abnormalities, epithelial-mesenchymal transition, and fibroblast activation.6-8 Furthermore, many studies have shown that the distribution of IPF-associated LC is localized to the peripheral areas of the lower lobes that are associated with honeycomb lung. These are the areas where fibrosis is predominant, suggesting a histological association between IPF and LC.9 Therefore, progressive bronchiolar proliferation in the fibrotic area could be a precursor of LC in IPF patients.7, 9 These reports suggest that IPF might be a precancerous lesion of LC. Therefore, in this study we investigated the molecular mechanisms of IPF-associated LC to clarify the impact of somatic mutation accumulation in the IPF region in tumorigenesis.
In the IPF region reported here, we detected five genetic alterations that encoded the receptor tyrosine kinases (KDR, EPHA5, and ERBB2) and the tumor suppresser genes (APC and CREBBP).10-14 In the LC region, we detected four genetic alterations that encoded the receptor tyrosine kinase (EPHA5), receptor-like protein (PKHD1), cell cycle regulator (RB1), and a substrate adaptor protein that senses oxidative stress (KEAP1).15-17 The variant allele frequencies at the IPF region were extremely low compared to those of LC. In our study, almost all somatic mutations did not overlap between the IPF and LC regions. Although the EPHA5 mutation was common in both tissues, it was considered to have less pathological significance because the information contained in the ClinVar database shows that it is probably benign. Another recent report showed that somatic alterations were not frequently shared between LC and the corresponding IPF tissue.18 Therefore, IPF-associated LC may be unlikely to be caused by a multistep accumulation of somatic alterations in the epithelium of honeycomb lung. There may be heterogeneity in the formation of IPF or LC in the lung tissue of IPF-associated LC. Furthermore, the RB1 mutation was considered to have a strong impact on the development of LC because it has been reported as a pathogenic mutation in the ClinVar database.
In the present report, the LMD techniques was used for the first time, to assess the targeted honeycomb lung epithelial cells from lung biopsy sections to investigate genetic mutations. LMD techniques are useful for genomic analyses in heterogeneous tissue samples.19 The present results of genetic mutations in each IPF and LC region, using LMD, were more precise than before.
Finally, our study has some limitations. The EPHA5 mutation might be a germline mutation. Since our case was not analyzed in pairs with normal tissues (e.g., blood), this possibility was not completely ruled out. In addition, variants could not be validated by different methods because of tissue limitations. Furthermore, it is necessary to investigate more cases with comprehensive NGS in the future since we investigated only one case with limited targeted NGS.
In conclusion, this is the first study to report that somatic mutations between the epithelium of IPF and LC regions were almost not overlapping by targeted NGS analysis using the LMD technique. Therefore, it is possible that IPF may not be a precancerous lesion of LC genetically, although pathogenic similarities in both have been reported.
ACKNOWLEDGMENTS
We wish to thank DNA Chip Research Inc. for technical assistance.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.