EEF1A2 and ERN2 could potentially discriminate metastatic status of mediastinal lymph node in lung adenocarcinomas harboring EGFR 19Del/L858R mutations
Abstract
Background
Early data has indicated that EGFR 19Del mutation and EGFR L585R mutation are two different types of non-small cell lung cancer (NSCLC). However, how the different molecular mechanisms participate in the process of mediastinal lymph node metastasis (MLNM) in lung adenocarcinoma (LA) harboring EGFR 19Del and EGFR L858R mutation remains unknown. We thus explored the genes responsible for MLNM in LA with EGFR 19Del or L858R mutation.
Methods
We performed transcriptome sequencing and bioinformatics analysis from 10 patients with LA resection specimens of primary tumors. Quantitative reverse transcription-polymerase chain reaction was used to validate gene expressions.
Results
There were 69 mRNAs upregulated and 100 mRNAs downregulated in five samples with MLNM compared with samples without MLN metastasis. EEF1A2 and ERN2 were observed exhibiting different expression patterns in EGFR 19Del and EGFR L858R samples with MLNM. In samples harboring EGFR 19Del mutation, the expression of EEF1A2 gene in samples with MLNM was significantly lower compared with samples without MLN metastasis, and in samples with EGFR L858R, it was significantly higher in samples with MLNM. The expression pattern of ERN2 was opposite to EEF1A2. In addition, several other genes including SLC6A11, IGHV3-48, IGHV3-43, DUSP9, and HOXA9 were also shown to be associated with invasion and metastasis and exhibited an expression pattern similar to EEF1A2 and ERN2 in EGRF 19Del and L858R mutation tumors.
Conclusions
EEF1A2 and ERN2 were for the first time observed exhibiting distinct expression patterns in MLNM in lung adenocarcinomas harboring EGFR 19Del and EGFR L858R mutation by interindividual DEGs analysis.
Key points
Significant findings of the study
In our study, we focused on the mechanisms of metastasis and invasion that different EGFR mutations conferred and identified two critical genes separately involved in this process in EGFR 19Del and L858R mutation tumors.
What this study adds
Our findings not only reinforced theoretical foundations that the EGFR 19Del and L858R mutation tumors should be considered as two kinds of diseases, but also laid the fundamentals for precise determination of the mediastinal lymph node radiation field and improvement of clinical outcome.
Introduction
Primary bronchogenic carcinoma arising in the lungs is a leading cause of death associated with cancer and emerges as a severe risk for human health.1 Radiotherapy is one of the major treatments in comprehensive therapeutics of lung cancer; however, there is about 5%–15% prevalence of radiation-induced lung injury (RILI) in patients treated with definitive chest radiotherapy, which usually can lead to serious clinical complications.2, 3 In order to avoid RILI and improve clinical outcomes, it is crucial to precisely predict mediastinal lymph node metastasis (MLNM) with potential molecular signature.4
Over the past decade, growing enthusiasm had been focused on identification of differentially expressed genes (DEGs) to predict mediastinal lymph node metastasis in lung cancer. Recently, evidence has shown that DEGs such as ADAM metallopeptidase domain 28 (ADAM28), C-X-C motif chemokine receptor four are closely associated with mediastinal lymph node metastasis in lung cancer and could be used as predictive biomarkers.5, 6
The RNA sequencing results revealed by Grigoroiu et al.7 demonstrated that there was significant overlap (only one gene) in optimized DEGs for prediction of MLNM between squamous cell carcinomas (23 genes) and adenocarcinoma (43 genes) in lung cancer, which suggests rather distinct mechanisms are involved in the development of MLNM in different histological subtypes. These preliminary studies have highlighted identification and functional investigation of DEGs, which can be utilized as a predictive biomarker of MLNM, could improve precise target delineation of MLN and clinical outcome of radiotherapy.
Although ATP-competing tyrosine kinase inhibitors (TKIs) have been widely used in clinical practice for advanced non-small cell lung cancer (NSCLC) with sensitive mutations in epidermal growth factor receptor (EGFR),8, 9 the combination of TKIs and radiotherapy treatment had a bottleneck for clinical utilization, mainly due to RILI.10, 11 Furthermore, the results from the LUX-Lung 3 and 6 13 that patients harboring EGFR 19Del mutation unequivocally benefit from treatment with TKIs compared with chemotherapy (mOS: 31.7 months vs. 20.7 months, P = 0.0001), and in contrast, the patients with EGFR L858R mutation do not completely benefit from EGFR TKIs (mOS: 22.1 months vs. 26.9 months, P = 0.160), strongly suggests that tumors driven by EGFR 19Del mutation are different from those of L858R mutation with regard to their biological significance.
Meanwhile, a difference in benefit from chemotherapies has also been observed in patients with different EGFR mutation status. Studies have reported that patients harboring EGFR 19Del mutation could be more responsive to chemotherapies than those with L858R mutations.14-16 In a previously published meta-analysis, Wu et al.17 demonstrated that EGFR 19Del mutation and EGFR L585R mutation are two different types of NSCLC and EGFR 19Del mutation can be utilized as both predictive and prognostic biomarkers. Accordingly, it is warranted to investigate the underlying mechanisms of the biological characteristics, especially MLNM, in these two different types of tumors.
With the intention of identification of the specific predictive biomarkers for MLNM in NSCLC patients with EGFR sensitive mutations thereby reducing chest targeted volume of radiotherapy and incidence of RILI, 10 specimens (five with MLNM and five without MLNM) from patients with lung adenocarcinoma harboring EGFR sensitive mutations were subjected to transcriptome sequencing and bioinformatic analysis to identify the genes responsible for MLNM. Based on RNA-Seq results, an analysis was performed on the distinct molecular mechanisms of MLNM in EGFR 19Del and L858R subtypes. These investigations should be useful to facilitate a rational combination of EGFR-TKI treatment and radiotherapy and provide theoretical fundamentals for further clinical practice of personalized therapeutics.
Methods
Patients and samples
A total of 10 patients with lung adenocarcinoma who underwent resection of primary lesions and lymphadenectomy of mediastinal lymph nodes between January 2014 and December 2014 were enrolled into this study. The median age was 66 (range: 51–73). Clinical recorded information such as age, gender, height, weight, smoking history, childbearing history, family medical history and clinicopathologic characteristics such as stage, histology, ECOG performance scores, EGFR mutations, surgery and MLNM were collected. Smoking history was categorized into three groups: never-smokers: ≤100 cigarette/lifetime or never smoking, ever-smokers: >100 cigarette/lifetime and abstaining from smoking >1 year, smokers: >100 cigarette/lifetime and abstaining from smoking <1 year or being still smoking. A total of 10 specimens from surgically-resected primary cancer were frozen and kept in liquid nitrogen. All patients provided their written informed consent for molecular analysis. The ethical approval was obtained from Daping Hospital Ethics Committee.
RNA quantification and qualification
RNA degradation and contamination were monitored on 1.5% agarose gels. RNA was extracted from samples using the NanoDrop 2000 (Thermo, CA, USA). RNA concentration and integrity were detected by the RNA Nano 6000 assay kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
Library preparation for transcriptome sequencing
A total amount of 5 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext Ultr RNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer's recommendations and index codes were added to attribute sequences to each sample. Poly-(A) mRNA was isolated from total RNA using Oligo-(dT) magnetic beads and then fragmented in fragmentation buffer. The first-strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-). Second-strand cDNA was synthesized using buffer, dNTPs, RNaseH, and DNA polymerase I. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3′ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure were ligated to prepare for hybridization. In order to select cDNA fragments of preferentially 250–300 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). Then, 3 μL USER Enzyme (NEB, USA) was used with size-selected, adaptor-ligated cDNA at 37°C for 15 minutes. PCR was performed with Q5 Hot Start HiFi DNA polymerase, Universal PCR primers,and Index (X) Primer. At last, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system. All the raw data of transcriptome sequencing are available via the following website: NCBI FTP: Address: ftp-private.ncbi.nlm.nih.gov (Table S5).
Clustering and sequencing
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v4-cBot-HS (Illumia) according to the manufacturer's instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2500 platform and paired-end reads were generated.
Data analysis
Clean data (clean reads) were obtained by removing reads containing adapter, reads containing ploy-N and low quality reads from raw data. Then, Q20, Q30 and GC content the clean data were assessed. The clean high quality data was the basis for all the following analyses.
Reads mapping to the reference genome
In order to conduct RNA-Seq experiments, reference genome and gene model annotation files were obtained directly from the genome website. Index of the reference genome was built using Bowtie v2.2.3 and reads were blasted to the reference genome using TopHat v2.0.12. We selected TopHat as the mapping tool as TopHat can generate a database of splice junctions based on the gene model annotation file and thus a better mapping result than other nonsplice mapping tools.
Quantification of gene expression level
The expression level of each gene was conducted based on RNA-Seq was measured as numbers of reads per kilobase of exon region in a gene per million mapped reads (RPKM). RPKM, expected number of fragments per kilobase of transcript sequence per millions of base pairs sequenced, considers the effect of sequencing depth and gene length for the reads count at the same time, and is currently the most commonly used method for estimating gene expression levels.18
Differential expression analysis
Prior to differential gene expression analysis, for each sequenced library, the read counts were adjusted by the edgeR program package through one scaling normalized factor. Differential expression analysis of two samples was performed using the DEGseq (2010) R package. P-value was adjusted using q value.19 FDR <0.05 and |log2Ratio| ≥ 1 found by DESeq was set as the threshold for significantly differential expression.
GO and KEGG enrichment analysis of differentially expressed genes
In order to analyze gene ontology (GO) enrichment of differentially expressed genes (DEG), we used the top GO software. GO terms with corrected P-value less than 0.05 were considered significantly enriched by differentially expressed genes.
Kyoto Encylopedia of Genes and Genomes (KEGG) is a database resource for understanding high-level functions and utilities of the biological system, such as the cell, the organism and the ecosystem, from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies (http://www. genome.jp/kegg/). We used R software to test the statistical enrichment of differential expression genes in KEGG pathways.
Quantitative RT-PCR
The expression levels of the gene ERN2 were validated by real-time RT-PCR. The cDNA was synthesized with RevertAid Fist Strand cDNA Synthesis Kit (ThermoFisher, United States, Category Number: #K1622). Primers for ERN2 were 5′-TCGAAGGACCAATGTACGTCA-3′ and 5′-GGATGGTGAATGGCAGTTTC AT-3′ and purchased from Bioligo Inc. (Shanghai China). Real time quantitative RT-PCR was performed with the SYBR Green PCR method using Fast SYBR Green Master Mix (ThermoFisher, United States, Category Number: 4385610) with CFX96 Real-Time System (Bio-Rad, United States). GAPDH was used as endogenous normalization controls. 2-△△Ct method was used to calculate the expression levels. Real-time PCR was performed in duplicate in three dependent experiments.
Results
Clinical characteristics
The clinical characteristics of the 10 patients enrolled into the study with lung adenocarcinoma are shown in Table 1. All patients in the study were female. This cohort comprised five cases with stage I, three cases with stage II, and two cases with stage III. All patients were ECGO ≤2 and never-smokers. EGFR 19Del mutation was detected in two cases (20%) and EGFR L858R mutation in eight cases (80%). Half of the population had MLNM and the another half did not.
| Characteristics | N (%) | N (%) |
|---|---|---|
| With MLN metastases | Without MLN metastases | |
| Total No. of patients | 5 | 5 |
| Median age (range) | 67 | 62 |
| Gender | ||
| Male | 0 | 0 |
| Female | 5 | 5 |
| ECOG PS | ||
| 0 | 0 | 1 |
| 1 | 5 | 4 |
| 2 | 0 | 0 |
| Tumor stage | ||
| I | 0 | 5 |
| II | 3 | 0 |
| III | 2 | 0 |
| IV | 0 | 0 |
| Histological type | ||
| Adenocarcinoma | 5 | 5 |
| Squamous cell carcinoma | 0 | 0 |
| Smoking status | ||
| Never-smokers | 5 | 5 |
| Ever-smokers | 0 | 0 |
| Smoking | 0 | 0 |
| EGFR | ||
| 19Del | 1 | 1 |
| L858R | 4 | 4 |
| MLN | ||
| Yes | 5 | 0 |
| No | 0 | 5 |
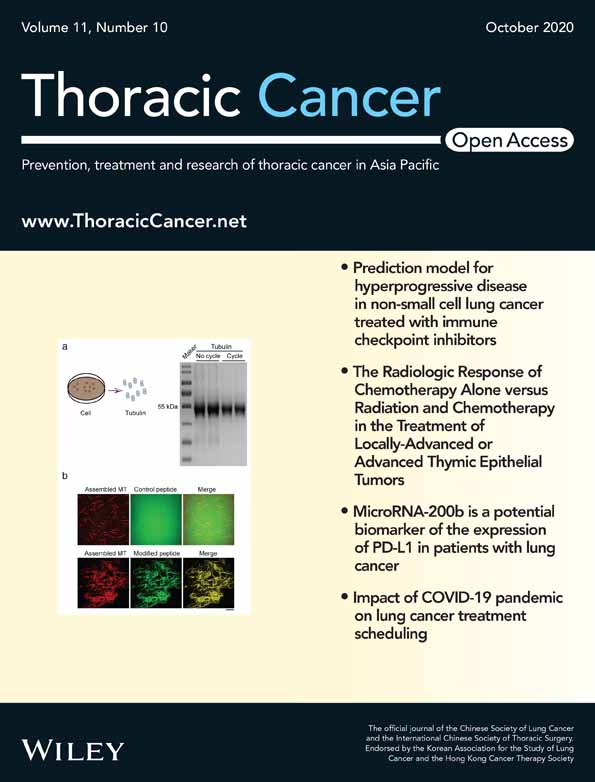
Transcriptome sequencing analysis revealed 169 DEGs
Collectively, 78 360 246 clean reads with Q20 > 98% were obtained from RNA-Seq in 10 samples. There were 69 mRNAs upregulated and 100 mRNAs downregulated in five samples with MLN metastasis compared with other five samples without MLN metastasis (FDR <0.05 and |log2Ratio| ≥ 1) (Table S1). The top 10 genes identified are listed in Table 2. Unsupervised clustering based on these 169 genes are shown in Fig 1. However, these 169 genes failed to distinguish the MLN metastasis samples from those without MLN metastasis because of interindividual gene expression diversity.
| Gene | log2 (Fold_change) | P-value | Regulation |
|---|---|---|---|
| KRT6A | −6.196 089 948 | 9.60E-09 | Down |
| SLC6A4 | −5.837 933 566 | 3.37E-05 | Down |
| FGG | −5.722 328 829 | 3.39E-08 | Down |
| LGALS4 | −4.190 533 502 | 1.24E-06 | Down |
| TMEM100 | −4.155 788 402 | 7.72E-07 | Down |
| CLDN18 | −3.986 126 264 | 1.34E-11 | Down |
| KRT14 | −3.961 357 307 | 4.22E-05 | Down |
| ITGB3 | −3.147 825 383 | 5.05E-05 | Down |
| AGER | −3.141 408 264 | 2.62E-07 | Down |
| BMP2 | −3.048456914 | 3.23E-05 | Down |
| KLK7 | 4.957 071 959 | 9.63E-05 | Up |
| KLK6 | 5.135 153 885 | 5.98E-05 | Up |
| ENSG00000214546.3 | 5.584 308 702 | 0.00014412 | Up |
| PRR4 | 6.874 154 524 | 4.71E-62 | Up |
| PDIA2 | 7.191 046 461 | 2.71E-22 | Up |
| PRB2 | 8.812 552 024 | 0.000405085 | Up |
| TRIM48 | 9.001887299 | 1.02E-06 | Up |
| PRB3 | 9.091590046 | 1.27E-33 | Up |
| PRB1 | 10.183 904 16 | 4.40E-08 | Up |
| PRB4 | 12.112 029 86 | 1.41E-37 | Up |
Interindividual DEGs analysis discovered two critical DEGs
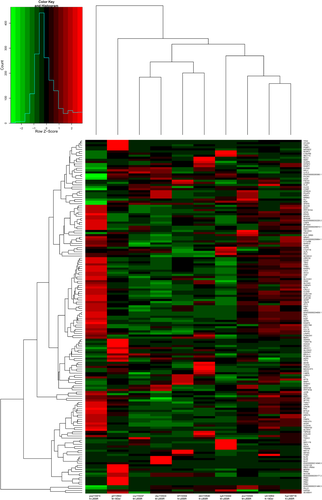
In order to avoid difficulties caused by heterogeneity of gene expression between samples, eight samples harboring EGFR L858R mutation were preprocessed with consistency analysis and four paired samples were extracted. The other two samples with EGFR 19Del mutation were matched (Table 3). The five paired samples were then subjected to individual DEG analysis with criteria of FDR≤0.05 and absolute value of log2Ratio ≥ 1 to identify DEGs (Fig 2). This analysis totally identified 1011 significantly differently expressed genes. Amongst these genes, EEF1A2 (ENSG00000101210, eukaryotic translation elongation factor 1 alpha 2) and ERN2 (ENSG00000134398, endoplasmic reticulum to nucleus signaling 2) were, for the first time, observed exhibiting different expression patterns in EGFR 19Del and EGFR L858R samples with MLN metastasis. Namely, in samples harboring EGFR 19Del mutation, the expression of EEF1A2 gene in the sample with MLN metastasis was significantly lower compared with the sample without MLN metastasis, and in samples with EGFR L858R mutation, it was significantly higher in samples with MLN metastasis. The expression pattern of ERN2 in EGFR 19Del mutation and EGFR L858R mutation samples was opposite to EEF1A2 (Table S2).
| Group | Total | Up | Down |
|---|---|---|---|
| ctm110540-vs.-cby110324 | 285 | 69 | 216 |
| hyz140716-vs.-syh110444 | 207 | 82 | 125 |
| trf110334-vs.-cxy110247 | 291 | 165 | 126 |
| wh14054-vs.-jzl110502 | 212 | 94 | 118 |
| ysq110474-vs.-zxs110343 | 319 | 245 | 74 |
 ) and (
) and ( )
)WGCNA identified functionally associated genes potentially responsible for MLN metastasis
Weighted gene co-expression network analysis (WGCNA) demonstrated that several genes such as MAP7D2, SMOC1, NOMO3, MEGF10, SLC5A3, NAT8L, CLDN9, BIRC5 and DLX6-AS1 were co-expressed with EEF1A2 and clustered with EEF1A2 as one functional module (Table S3). Like EEF1A2, these genes were upregulated in samples with MLN metastasis with EGFR L858R mutation and downregulated in samples with MLN metastasis with EGFR 19Del mutation compared with corresponding samples without MLN metastasis. Other genes including PI3, SERPINA1, SP5, CACNA1F, ELF5, CTSE, MUC7, UGT2B7, AGXT, RP11-366L20.4 and CTD-2196E14.8 were clustered with ERN2 and classified as one functional module (Table S4). The functions in the majority of these genes are relevant to translocases (Fig 3).
Genes involved in DNA damage repair and immunodeficiency collaboratively participate in MLN metastasis
With the aim of further revealing the underlying mechanisms separately mediating MLN metastasis in EGFR 19Del and L858R mutation tumors in more detail, the paired sample wanghong14054-vs.-jiangzhilan110502 was analyzed to identify more relevant genes involved in MLN metastasis. In addition to EEF1A2 and ERN2, other genes including SLC6A11, IGHV3-48, IGHV3-43, DUSP9 and HOXA9 were shown to be associated with invasion and metastasis and exhibited expression patterns similar to EEF1A2 and ERN2 in EGRF 19Del and EGFR L858R mutation tumors. Functional enrichment analysis with KEGG found that these genes were all associated with transcriptional misregulation in cancer. Moreover, IGHV3-48 and IGHV-43 also participated in primary immunodeficiency. Accordingly, we speculated that different genes, especially in DNA damage repair and the autoimmune system participate in MLN metastasis in EGFR 19Del and L858R mutation tumors (Fig 4).
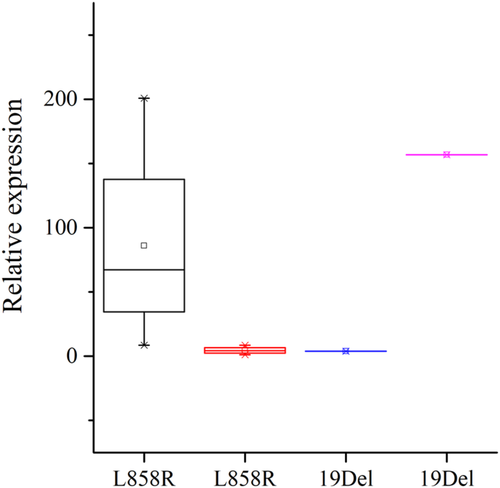
Quantitative RT-PCR verifies RNA-seq results
Quantitative TR-PCR was used to verify ERN2 expression in the same 10 samples. Owing to the limited sample size, there was just a trend that the expression of ERN2 was lower in EGFR L858R mutation and higher in EGFR 19Del mutation samples with NLM metastasis compared with corresponding samples without NLM metastasis (Fig 5).
 ) Non- MLNM (
) Non- MLNM ( ) MLNM (
) MLNM ( ) Non MLNM (
) Non MLNM ( ) MLNM
) MLNMDiscussion
EGFR-TKIs combined with radiotherapy has been shown to be a promising comprehensive therapy for advanced or metastatic NSCLC, dramatically improving individualized treatments.8, 9 However, this therapeutic strategy is limited in clinical practice mainly due to the severe risk of RILI. The ability to precisely anticipate MLN metastasis and reduce the field size of mediastinal irradiation in order to reduce the incidence of RILI for utilization of a combination of EGFR TKIs and radiotherapy is therefore urgently required.
In an effort to identify specific biological predictors for MLNM, DEG analysis revealed that 69 genes including AQP5 were significantly upregulated and 100 genes such as KRT6A were significantly downregulated in specimens from patients with MLNM compared with that without MLNM. Taking into consideration the biological difference between EGFR 19Del mutations and EGFR L858R mutation, EEF1A2 and ERN2 were observed exhibiting distinct expressional patterns by interindividual DEG analysis for the first time.
EEF1A2 is located at 20q13.3 and is a housekeeping gene. The protein eEF1A2 encoded by this gene can regulate interaction between ribosome and mRNA codes after combination with tRNA, which has an important role for elongation of protein translation.20 Some reviews summarized the recent findings on eEF1A proteins indicating that eEF1A protein plays a critical role in numerous human diseases through enhancement of oncogenesis, blockade of apoptosis, and increased viral pathogenesis.21 Several studies have shown that expression of EEF1A2 is abnormally upregulated in lung cancer and others type of carcinomas such as liver cancer,22 cervical cancer,23 pancreatic ductal adenocarcinoma24 and breast cancer.25 The ERN2 gene is located in 16p12.2 and has an important role in signaling transduction from endoplasmic reticulum to nucleus.26 ERN2 also activates serine/threonine-protein kinase and participates in the response to EGFR-TKIs.27, 28 Our study observed that the expression of EEF1A2 was downregulated in patients with MLNM harboring EGFR 19Del mutations compared with those patients without MLNM, and conversely upregulated in patients with MLNM harboring EGFR L858R mutations, whereas the expression of ERN2 was upregulated in patients with EGFR 19Del mutations and downregulated in those with EGFR L858R mutations. The expression of ERN2 was verified with quantitative RT-PCR in these 10 patients and found to be consistent with the results from transcriptome sequencing.
In addition to EEF1A2, WGCNA revealed that several other genes including MAP7D2, SMOC1, NOMO3, MEGF10, SLC5A3, NAT8L, CLDN9, BIRC5 and DLX6-AS1 were all downregulated in patients with EGFR 19Del mutation and upregulated in those with EGFR L858R mutation which suggested genes in the same cluster participate together in a biological function. Genes that clustered with ERN2 included PI3, SERPINA1, SP5, CACNA1F, ELF5, CTSE, MUC7, UGT2B7, AGXT, RP11-366L20.4, and CTD-2196E14.8. The function in the majority of these genes is to transport enzymes.
In order to further uncover the difference in mechanisms of MLN metastasis between EGFR 19Del mutation and L858R mutation, individual DEG analysis was conducted in pair of specimens of wanghong14054-vs.-jiangzhilan110502. The results revealed that besides EEF1A2 and ERN2, several other genes such as SLC6A11, IGHV3-48, IGHV3-43, DUSP9 and HOXA9 were also involved in invasion and migration exhibiting low expression in samples with EGFR 19Del mutations and high expression in samples with EGFR L858R mutations. These genes function as regulators involved in transcriptional misregulation in cancer.29-33 IGHV3-48 and IGHV3-43 also participate in the pathways of primary immunodeficiency.30, 31 Altogether, these results indicated many genes are associated with DNA damage repair and regulation of immune system cooperatively regulate the MLNM with different mechanisms between EGFR 19Del and EGFR L858R mutations.
The results obtained above strongly suggested that different mechanisms are involved in MLNM in these two subtypes of lung adenocarcinoma, which is consistent with a recently proposed concept that lung adenocarcinoma with EGFR 19Del mutation is quite different from that with EGFR L858R mutation. Many studies have reached this conclusion. First, although lung adenocarcinoma cell lines transfected with 19Del or L858R mutation EGFR are both more or less sensitive to gefitinib and the expression of p-AKT and p-ERK are inhibited in these two transfected cell lines, the inhibitory effects in cells with 19Del mutation is stronger than that in L858R mutation.34 Cui et al. has shown that the autophosphorylation site of EGFR with L858R mutation is different from that with 19Del mutation, and the tyrosine residues of the 845 code in EGFR with L858R mutation is highly phosphorylated which results in different downstream signaling pathways.35 Furthermore, the equilibrium constant Ki of interaction between erlotinib and EGFR with 19Del mutation is far lower than that of EGFR with L858R mutation.36 Wang et al. revealed that EGFR protein with 19Del mutation exhibits lower degradation rate than EGFR protein with L858R mutation because of lower interaction with c-Cbl.37 Second, from a clinical point of view, in a randomized controlled phase III clinical trial (EURTAC study) 16 that compared first-line EGFR TKI treatment with chemotherapy in advanced NSCLC, patients with different EGFR mutation types had different benefit extents from EGFR TKI. Subset analysis demonstrated that compared with the chemotherapy, patients with EGFR L858R mutation had a moderately reduced risk of progression (HR = 0.55, P = 0.0539), whereas the patients with EGFR 19Del mutation had a rather stronger decrease in risk of progression (HR = 0.30, P < 0.0001). Third, a meta-analysis (n = 1489) including seven large phase III randomized controlled clinical trials (IPASS, WJTOG3405, EUTRAC, OPTIMAL, LUX-LUNG3, LUX-LUNG6, NEJ002, ARCHER1050, FLAUR) suggested that patients with advanced NSCLC harboring EGFR 19Del mutation could benefit more from treatment with EGFR TKIs than patients with EGFR L858R mutations (HR19/21 = 0.587, 95% CI: 0.376–0.917, P = 0.02).38-40 Zhang et al.41 also achieved the same conclusion through their meta-analysis based on 13 clinical trials. The results from a pooled analysis combining patients from LUX-LUNG 3 and LUX-LUNG 6 also showed that patients with EGFR 19Del mutation exhibited clear benefits from afatinib treatment with respect to OS (HR: TKI vs. chemotherapy = 0.59, 95% CI: 0.45–0.77, P = 0.0001); in contrast, no benefits from afatinib treatment were observed in patients with EGFR L858R mutation (HR: TKI vs. chemotherapy = 1.25, 95% CI: 0.92–1.71, P = 0.16). These results could be attributed to fact that patients harboring EGFR 19Del mutation had a better response to EGFR TKI treatment than those with EGFR L858R mutation.12, 13
Recently, growing evidence has suggested that it is important to understand the function of different hotspot mutations in the same driver gene which can cause different biological consequences such as carcinogenesis, invasions, resistance to therapeutics and prognosis. In our study, we focused on the mechanisms of metastasis and invasion that different EGFR mutations conferred and identified two critical genes separately involved in this process in EGFR 19Del and L858R mutation tumors. Despite the fact that our study was limited to 10 patients, our findings not only reinforced the theoretical foundations that EGFR 19Del and L858R mutation tumors should be considered as two kinds of diseases, but also laid the foundations for the precise determination of mediastinal lymph node radiation fields and improvement of clinical outcome after radiotherapy.
Acknowledgments
The authors thank Mr Xian-Feng Lu and Miss Mao-Jun Liao for their kind and excellent technical assistance. This study was sponsored by the Chongqing Science and Health Combined Medical Research Project (No. 2019MSXM024) to Lei Xia.
Disclosure
The authors declare that there are no competing interests.