Evaluating restriction enzyme selection for reduced representation sequencing in conservation genomics
Carlos Carreras, Marta Pascual, and Cinta Pegueroles jointly supervised this work.
Abstract
Conservation genomic studies in non-model organisms generally rely on reduced representation sequencing techniques based on restriction enzymes to identify population structure as well as candidate loci for local adaptation. While the expectation is that the reduced representation of the genome is randomly distributed, the proportion of the genome sampled might depend on the GC content of the recognition site of the restriction enzyme used. Here, we evaluated the distribution and functional composition of loci obtained after a reduced representation approach using Genotyping-by-Sequencing (GBS). To do so, we compared experimental data from two endemic fish species (Symphodus ocellatus and Symphodus tinca, EcoT22I enzyme) and two ecosystem engineer sea urchins (Paracentrotus lividus and Arbacia lixula, ApeKI enzyme). In brief, we mapped the sequenced loci to the phylogenetically closest reference genome available (Labrus bergylta in the fish and Strongylocentrotus purpuratus in the sea urchin datasets), classified them as exonic, intronic and intergenic, and studied their function by using Gene Ontology (GO) terms. We also simulated the effect of using both enzymes in the two reference genomes. In both simulated and experimental data, we detected an enrichment towards exonic or intergenic regions depending on the restriction enzyme used and failed to detect differences between total loci and candidate loci for adaptation in the empirical dataset. Most of the functions assigned to the mapped loci were shared between the four species and involved a myriad of general functions. Our results highlight the importance of restriction enzyme selection and the need for high-quality annotated genomes in conservation genomic studies.
1 INTRODUCTION
We are facing the sixth mass extinction on Earth, with an accelerated global loss of biodiversity (IPBES, 2019). In the last decades, genetics has made it possible to delve into important processes of interest for conservation such as the level of inbreeding or gene flow between or within populations (Ouborg et al., 2010). However, there are still unresolved questions, and this is where conservation genomics plays a critical role. While conservation genetics is based on a reduced number of loci, conservation genomics is based on thousands of genome-wide loci. Genomics can help biodiversity conservation (Theissinger et al., 2023) and improve our understanding of evolution and adaptation in the marine environment, even in non-model organisms (Nielsen et al., 2009). Genome-wide loci allow the detection of population adaptation patterns elusive with fewer loci (Bradbury et al., 2015). Reduced representation techniques are used in population genomics to increase locus coverage to ensure reliable genotyping of many individuals at a lower sequencing cost, without compromising their genetic differentiation (Galià-Camps et al., 2022, 2023). Reduced representation of the genome by enzymatic digestion and high-throughput genotyping techniques can be applied even in species without a reference genome (Andrews et al., 2016). Among these techniques, Genotyping-by-Sequencing (GBS) is a simple system for building libraries and massively parallel sequencing, to discover from hundreds to thousands of genome-wide loci (Elshire et al., 2011). When using reduced representation techniques, it is assumed that the sequenced fraction is representative of the whole genome; however, the reduction of the genome might depend on the recognition site of the restriction enzyme used. Consequently, the genomic composition of the candidate loci (the proportion of loci in exonic, intronic or intergenic regions) and their functional composition (the biological functions assigned, for instance, Gene Ontology [GO] terms) could be influenced by restriction enzymes, resulting in potential biases. In fact, previous studies showed that the distribution of loci obtained with different restriction enzymes using nucleotide distributions (Herrera et al., 2015) or simulated data (Rivera-Colón et al., 2021) are highly variable among taxonomic groups. Thus, we need a better understanding of the extent restriction enzyme selection influences genomic studies.
Population genomic studies published in different taxa are of particular interest since they allow the evaluation of the effect of the genomic technique used (Carreras et al., 2020, 2021; Torrado et al., 2020). Carreras et al. studied the genetic structure of the two species of sea urchins cohabiting in the Mediterranean Sea: the edible sea urchin Paracentrotus lividus (Carreras et al., 2020) and the black sea urchin Arbacia lixula (Carreras et al., 2021). Sea urchins are important engineers of infralittoral benthic communities, playing a key ecological role in controlling the structure of communities through grazing activity (Agnetta et al., 2015; Carreras et al., 2020; Palacín et al., 1998; Wangensteen et al., 2011). While P. lividus is mainly herbivorous, A. lixula has a tendency from omnivory to carnivory (Agnetta et al., 2013). Even though the two sea urchins have a role in the formation of barren patches (Bulleri, 2013; Bulleri et al., 1999), some studies show that A. lixula has a role in maintaining them (Bonaviri et al., 2011; Bulleri et al., 1999; Guidetti & Dulcić, 2007). Importantly, both species are facing the effects of global warming. The black sea urchin, A. lixula, is a thermophilic species (Pérez-Portela et al., 2019; Wangensteen et al., 2012) contrary to the purple sea urchin, Paracentrotus lividus, that prefers cold waters. During the past decades, several populations of P. lividus have been declining, and some of them even collapsed mainly due to high commercial interest (Yeruham et al., 2015). In addition, the current increase in the seawater temperature is expected to favour A. lixula, due to its more thermophilic biology and its phenotypic plasticity (Pérez-Portela et al., 2019). In both species, Carreras et al. (2020, 2021) identified some degree of population structure and candidate loci for adaptation associated with salinity and different temperature variables. They mapped the small fraction of loci under selection, showing that numerous candidate loci were located in exonic regions, suggesting that candidate loci could be enriched in exonic regions (Carreras et al., 2020, 2021).
The two endemic fishes from the Mediterranean Sea, Symphodus ocellatus and Symphodus tinca inhabit algal-covered rocky substrates and sea-grass beds like Posidonia oceanica (Macpherson et al., 2002). They are part of the Labridae family which represents a crucial connection of the trophic web in coastal environments (Shili et al., 2018). These two species are also considered supplementary fish cleaners, which help other fish (hosts) to be free of parasites (Zander & Sötje, 2002). While S. ocellatus is a microphagus predator (Macpherson et al., 2002), so it mainly feeds on Bryozoa, molluscs, and polychaetes (Quignard & Pras, 1986), S. tinca is a key species due to its abundance and generalist diet (Carreras et al., 2017), feeding on sea urchins, ophiuroids and molluscs (Quignard & Pras, 1986). In addition, S. ocellatus can be used as a fish model for ecological impact studies due to its high density and distribution (Levi et al., 2005). Torrado et al. (2020) found different levels of population structure across the Western Mediterranean in the two species, with higher population differentiation in S. ocellatus, in accordance with different dispersal distance distributions from backtracking modelling (Torrado et al., 2021). In both S. tinca and S. ocellatus, the authors found several candidate loci associated with temperature, productivity and turbulence variables. Contrary to sea urchins, most of the candidate loci identified in these two fish species were located in introns. In all four studies, loci were obtained by GBS using different restriction enzymes (ApeKI for sea urchins and EcoT22I for fish). The different genomic and functional composition of candidate loci in these four species is intriguing and may be attributed to differences in genomic composition between the two taxonomic groups, the use of different restriction enzymes or different selection processes mediating local adaptation. To evaluate the importance of these three processes in determining why candidate loci are mostly found in exons in sea urchins but introns in fish, and if there is an enrichment of these two genomic categories in candidate loci in these four species, it is necessary to compare the composition of the candidate loci to all genome-wide genotyped loci, which has not been addressed so far.
Here, we aim to test whether GBS data obtained using different restriction enzymes and species result in differential enrichment of genomic regions or/and functions, in all genome-wide and candidate loci. To do so, we analysed published data from four species (Carreras et al., 2020, 2021; Torrado et al., 2020), two endemic fish species with genomic libraries obtained with the EcoT22I enzyme (S. ocellatus and S. tinca), and two ecosystem-building sea urchins obtained with the ApeKI enzyme (P. lividus and A. lixula). By aligning the reference loci obtained from genotyping multiple individuals to the most nearby reference genome (Labrus bergylta in fish and Strongylocentrotus purpuratus in sea urchins), we classified loci as genic (distinguishing between exonic or intronic regions) and intergenic. Additionally, we simulated the use of the same two enzymes in both reference genomes and characterized the genomic category of the obtained markers. We evaluated the genomic composition of all annotated loci that mapped to unique positions and compared the genomic and functional composition of candidate loci and total loci, considering the different species and enzymes used.
2 MATERIALS AND METHODS
2.1 Species and data collection
We analysed published population genomics data of two fish (Actinopterygii) Symphodus ocellatus and Symphodus tinca) (Torrado et al., 2020) and two sea urchins (Echinoidea) Paracentrotus lividus (Carreras et al., 2020) and Arbacia lixula (Carreras et al., 2021). Genomic loci for the four species were obtained by GBS with EcoT22I for the two fish species, whose restriction site is (A | TGCA | T), where the bar identifies the cut sites generating sticky ends; and ApeKI for the two sea urchin species, whose restriction site is (G | CWG | C) where W can be either A or T.
In fish, the authors used the STACKS v1.47 software (Catchen et al., 2013) to identify haplotype loci and for genotyping, after trimming single-end sequenced reads to 59 bp (Torrado et al., 2020). Loci were obtained from 162 individuals of S. ocellatus and 141 of S. tinca collected in 6 and 5 different locations, respectively, along the Mediterranean coast of the Iberian Peninsula (Table S1). Several filtering steps were used to obtain the final dataset in both fish species (Torrado et al., 2020). In short, individual genotypes with a depth below 5 reads were not considered. Loci with a missingness value higher than 30% or with the major allele frequency equally or higher than 0.95 (i.e. monomorphic at that level) were removed. Finally, the loci in Hardy–Weinberg disequilibrium at more than 60% of the sampling sites were also eliminated from the final dataset. Overall, 3985 loci of S. ocellatus and 5284 loci of S. tinca were retained after filtering (Torrado et al., 2020). Candidate loci for adaptation were identified by obtaining individual-based data on four phenotypic variables (hatching date, planktonic larval duration, growth rate during planktonic larval duration, and settlement size) and three environmental variables (surface temperature, productivity and turbulence). Individual-based data were acquired from otolith readings. By using redundancy analysis (RDA) with environmental variables, genome-wide association studies (GWAS) with environmental and phenotypic variables, and outlier analysis, the authors of this study identified 7.3% and 3.2% of candidate loci to be under selection for S. ocellatus and S. tinca respectively (Table S1). In sea urchins, the authors used the GIbPSs toolkit (Hapke & Thiele, 2016) to de novo identify haplotype loci and for genotyping (Carreras et al., 2020, 2021). This software was used since it allowed working with paired-end sequences and did not require the same sequence length at different loci. Sequences were trimmed to 80 bp and posteriorly forward and reverse sequences of a paired-end assembled. Loci shorter than the read length were identified and only the forward read was kept resulting in shorter sequences. Thus, the size of the retained loci ranged from 35 to 152 bp. Several filtering steps were used to obtain the final dataset in both sea urchin species (Carreras et al., 2020, 2021). In short, individual genotypes with a depth below 5 reads were not considered. Loci potentially including an insertion/deletion, with more than two alleles per individual, or deeply sequenced were discarded. Finally, only loci present in at least 70% of the individuals were retained. The loci were obtained using 241 individuals of P. lividus and 240 of A. lixula collected in 11 different locations from the occidental and oriental Mediterranean basin and the eastern Atlantic coast (Carreras et al., 2020, 2021). Overall, 3730 loci of P. lividus and 5241 loci of A. lixula were retained after filtering (Carreras et al., 2020, 2021). Candidate loci for adaptation were identified by obtaining population-based environmental data (averaged from January 1993 to December 2016) at four temperature variables (mean, maximum, minimum and range) and four salinity variables (mean, maximum, minimum and range). By using RDA and outlier analyses, the authors of these studies identified 10.8% and 5.0% candidate loci to be under selection for P. lividus and A. lixula respectively (Table S1). For the four species, we obtained fasta files with the sequences of all the analysed haplotype loci using STACKS v1.47 in S. ocellatus and S. tinca, and GIbPSs toolkit in P. lividus and A. lixula.
2.2 Classification and data analysis of total and candidate loci
All the following analyses were performed for all the loci found in these studies (referred to as total loci) as well as for those loci candidates for adaptation found by the different approaches detailed in the previous section (referred to as candidate loci). To identify the genomic location of all the loci, we first mapped the sequences to the reference genome of the most closely related species using makeblastdb v2.10.1 followed by BLASTN searches that allow comparing distantly related homologous sequences (e-value ≤1e−4, outfmt = 6) and thus are appropriate to compare the studied loci to reference genomes of distant species. In fish, we used the genome of Labrus bergylta (BallGen_V1, assembly accession: GCF_900080235.1 including the fasta file and the GFF annotation) which diverged 28.2 MYA from Symphodus ocellatus and Symphodus tinca (http://www.timetree.org/ accessed in April 2022, Figure 1a). In sea urchins, we used the genome of Strongylocentrotus purpuratus as reference (Spur_5.0, assembly accession: GCF_000002235.5 including the fasta file and the GFF annotation) which diverged 183 MYA from A. lixula and 53.9 MYA from P. lividus (http://www.timetree.org/ accessed in April 2022, Figure 1a). We then classified sequences as uniquely mapped or mapping to multiple genomic locations, hereafter referred to as the “repeated class”. Finally, we characterized the uniquely mapped blast hits as genic (exonic and intronic), or intergenic using the in-house Python script classifyBlastOut.py (Figure 1b, script available in our GitHub repository, https://github.com/EvolutionaryGenetics-UB-CEAB/restrictionEnzimes.git). In brief, this script requires a file containing the coordinates of the blast hits mapped to unique genomic positions (in outfmt 6) and a GFF file with the features annotated in a given genome (must include at least genic and exonic information). By comparing coordinates, this script reports a file with the labels assigned to each blast hit, being genic (further distinguishing between exons, introns and providing gene IDs as stated in the GFF file) or intergenic. To calculate the percentage of exons, introns and intergenic regions in the reference genomes, we used the command genomecov with -d and -split options from BEDTools software (Quinlan & Hall, 2010). In order to use this software we first needed to convert the GFF files to BED12 format. The format conversion was done with two scripts from USCS utils (http://hgdownload.cse.ucsc.edu/admin/exe/linux.x86_64/), gtfToGenePred and genePredToBed. Count data were compiled in contingency tables. We checked for statistical differences in the loci classification, within and between species, using Fisher's exact tests implemented in R v4.1.0 (R Core Team, 2021).

2.3 In silico digestions of the two reference genomes
We generated simulated GBS data for the two reference genomes: Labrus bergylta (BallGen_V1, assembly accession: GCF_900080235.1) and Strongylocentrotus purpuratus (Spur_5.0, assembly accession: GCF_000002235.5) using the SimRAD package from R (Lepais & Weir, 2014). First, we in silico digested the two genomes using the ApeKI and EcoT22I enzymes independently. Then we selected fragments between 35 and 152 bp to match the sizes of the experimental data analysed in sea urchins. We used the same size selection with both enzymes and species to avoid methodological confounding effects. To evaluate their genomic composition, we first mapped the selected sequences to the corresponding reference genome using Hisat2 v2.2.1 software (Kim et al., 2019) because it is faster than BLASTN and the best tool when having a close reference genome. We then discarded those sequences that mapped to multiple positions in the genome using SAMtools view v0.1.19 (Danecek et al., 2021) and grep command (grep -P “(NH:i:1|^@)”). We finally identified the location of the uniquely mapped reads by comparing the filtered BAM files obtained with SAMtools and the GFF file for each species (see above) using BEDTools intersect (Quinlan & Hall, 2010). We checked for statistical differences in the loci classification between enzymes within species by performing Chi-squared tests implemented in R v4.1.0 (R Core Team, 2021).
2.4 Functional analysis
For the functional analysis, we assigned GO terms to the loci mapped to unique genomic regions using eggNOG-mapper v5 (Huerta-Cepas et al., 2019) in each species separately. To do so, we first made a list with the L. bergylta genes having significant unique blast hits with S. ocellatus and S. tinca, and a list with the S. purpuratus genes having significant unique blast hits with P. lividus and A. lixula. Using the GFF files from NCBI, we obtained the correspondence between Gene ID and Protein ID, and we generated a fasta file including the longest amino acid sequence for each identified gene. This file was used as input for the eggNOG-mapper using many-to-many orthology relationships within Metazoa. From the eggnog output file, we extracted the protein ID and the GO terms associated with them, and finally, we integrated the protein ID and GO terms with the genes name and locus ID of our species. Figure 1c shows a scheme of the pipeline used (https://github.com/EvolutionaryGenetics-UB-CEAB/restrictionEnzimes.git).
The analysis of the Gene Ontology (GO) terms was done using the online server Categorizer (https://www.animalgenome.org/bioinfo/tools/countgo/, accessed 07/2022). First, we classified the GO terms according to the root category they belonged to (biological process, molecular function and cellular component). Secondly, we classified the GO terms assigned to the biological process category, by using the 442 categories from the GO slims list from QuickGO (https://www.ebi.ac.uk/QuickGO/). GO slims are a list of selected terms, including cytoplasm organization, metabolic process, DNA replication, localization, signalling, cell death and circadian rhythm, which help summarize GO terms into broad high-level categories. A Venn diagram showing the presence of GO terms in the GO slims categories for each of the 4 species and their overlap was obtained using the ggvenn function from the ggplot2 package in R (Wickham, 2016). It is worth noting that the number of counts obtained from Categorizer can be higher than the total number of input GO terms since one GO term can belong to more than one category. The visualization of the shared GO terms was performed using the Revigo software (Supek et al., 2011).
3 RESULTS
3.1 Genomic characterization of total and candidate loci in fish and sea urchins
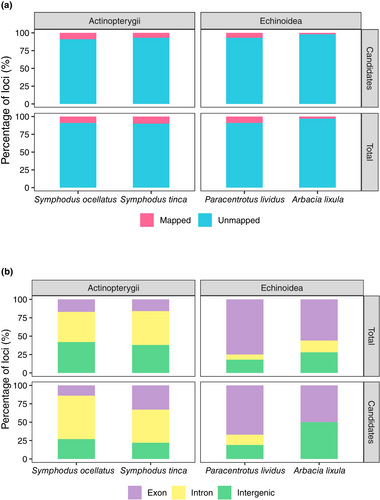
Overall, the frequency of loci mapping for each species to their respective reference genomes was low, with an average of less than 10% (Table 1 and Figure 2a). Within species, there were no significant differences in the mapping success for total and candidate loci as indicated by Fisher's exact tests (Table 2). We examined if there were significant differences between the two fish species (S. ocellatus vs. S. tinca) and between the two sea urchin species (P. lividus vs. A. lixula) for total and candidate loci mapping in their corresponding reference genomes (Table S1). The statistical tests showed no significant differences between S. ocellatus and S. tinca but significant differences between P. lividus and A. lixula, with a smaller frequency of mapped loci in the latter at both total and candidate loci (Tables 1 and 2). Differences between taxonomic groups at total loci (fish vs. sea urchins) were significant when considering the four species (Table 3). Knowing the significantly lower mapping success in A. lixula, which could bias the comparison between groups, we tested the differences between taxonomic groups excluding this species, resulting in non-significant differences (Table 3).
| Loci | Categories | Symphodus ocellatus | Symphodus tinca | Paracentrotus lividus | Arbacia lixula | ||||
|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | ||
| Total | Mapped | 423 | 10.6 | 512 | 9.7 | 342 | 9.2 | 174 | 3.3 |
| Unmapped | 3562 | 89.4 | 4772 | 90.3 | 3388 | 90.8 | 5067 | 96.7 | |
| Candidate | Mapped | 26 | 8.9 | 11 | 6.6 | 30 | 7.5 | 6 | 2.3 |
| Unmapped | 266 | 91.1 | 157 | 93.5 | 372 | 92.5 | 258 | 97.7 | |
| Total | Unique | 352 | 83.2 | 420 | 82.0 | 242 | 70.8 | 88 | 50.6 |
| Repeated | 71 | 16.8 | 92 | 18.0 | 100 | 29.2 | 86 | 49.4 | |
| Candidate | Unique | 22 | 84.6 | 9 | 81.8 | 21 | 70.0 | 2 | 33.3 |
| Repeated | 4 | 15.4 | 2 | 18.2 | 9 | 30.0 | 4 | 66.7 | |
| Total | Genic | 206 | 58.5 | 261 | 62.1 | 199 | 82.2 | 63 | 71.6 |
| Intergenic | 146 | 41.5 | 159 | 37.9 | 43 | 17.8 | 25 | 28.4 | |
| Candidate | Genic | 16 | 72.7 | 7 | 77.8 | 17 | 81.0 | 1 | 50.0 |
| Intergenic | 6 | 27.3 | 2 | 22.2 | 4 | 19.1 | 1 | 50.0 | |
| Total | Exonic | 61 | 29.6 | 66 | 25.3 | 183 | 92.0 | 49 | 77.8 |
| Intronic | 145 | 70.4 | 195 | 74.7 | 16 | 8.0 | 14 | 22.2 | |
| Candidate | Exonic | 3 | 18.8 | 3 | 42.9 | 14 | 82.4 | 1 | 100.0 |
| Intronic | 13 | 81.3 | 4 | 57.1 | 3 | 17.7 | 0 | 0.0 | |
- Note: The assigned categories of the loci were obtained by comparison to the corresponding reference genome, Labrus bergylta in fish and Strongylocentrotus purpuratus in sea urchins.
| Contrast | Total versus candidate | Total versus candidate | S. ocellatus versus S. tinca | P. lividus versus A. lixula | ||||
|---|---|---|---|---|---|---|---|---|
| SO | ST | PL | AL | Total | Candidate | Total | Candidate | |
| Mapped versus Unmapped | 0.428 | 0.229 | 0.312 | 0.476 | 0.144 | 0.477 | 0.000 | 0.004 |
| Unique versus Repeated | 1.000 | 1.000 | 1.000 | 0.682 | 0.666 | 1.000 | 0.008 | 0.161 |
| Exon versus Intron versus Intergenic | 0.288 | 0.310 | 0.335 | 1.000 | 0.349 | 0.492 | 0.001 | 0.585 |
- Note: For each analysis, we compared the number of loci falling in the different categories among total and candidate loci within species and between the two species within each taxonomic group. In bold are the significant values. Symphodus ocellatus (SO), Symphodus tinca (ST), Paracentrotus lividus (PL) and Arbacia lixula (AL).
| Contrast | All species | Without A. lixula | ||
|---|---|---|---|---|
| Chi-square | p-value | Chi-square | p-value | |
| Mapped versus Unmapped | 221.49 | <.0001 | 4.70 | .094 |
| Unique versus Repeated | 88.96 | <.0001 | 21.50 | <.0001 |
| Exon versus Intron versus Intergenic | 328.46 | <.0001 | 312.27 | <.0001 |
- Note: In bold are the significant values. The number of loci in each comparison can be found in Table 1.
The proportion of loci that mapped to unique and multiple (repeated) genomic locations did not differ significantly between total and candidate loci for any of the species (Table 1). In fish, most loci (>80%) mapped to unique positions without significant differences between or within species for total and candidate loci (Table 1). In the case of P. lividus and A. lixula, we found significant differences for the total loci, with a higher frequency of unique loci in P. lividus (Tables 1 and 2), but not for candidate loci, which may be due to the low number of candidate loci mapped in A. lixula. When we compared the frequency of unique loci between taxonomic groups, we obtained significant differences, with fish showing higher abundances independently of including or excluding A. lixula (Table 3).
We further classified the loci mapped to unique positions as being located in exonic, intronic or intergenic regions (Figure 2b). Overall, in the four species, we observed a majority of loci being in genic regions (exons and introns). However, the percentage of total loci that hit genic regions was higher in P. lividus and A. lixula (82% and 72% respectively) than in S. ocellatus and S. tinca (59% and 62% respectively), despite the similar percentage of genic regions in their respective reference genomes (35%, Table S2). In fish, most loci in genic regions mapped to introns, contrary to sea urchins, where most loci mapped to exonic regions, despite the fact that the percentage of exons was very similar in the two reference genomes (6.5%, Table S2). There were no significant differences, between total and candidate loci within each species, in the frequency of loci mapping in exonic regions (Table 2). We did not detect significant differences between Symphodus species in the abundance of genes mapping in exonic regions in total or candidate loci, but we detected significant differences between sea urchins, especially when analysing the total loci (Table 2). In addition, there were significant differences in exonic loci when comparing fish and sea urchins both considering and not considering A. lixula (Table 3).
3.2 Genomic composition of the two reference genomes in silico digestions
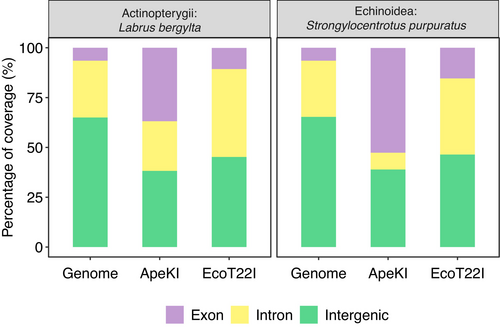
In order to evaluate the importance of the restriction enzyme when using reduced representation sequencing techniques, we generated in silico GBS data for the two reference genomes simulating their digestion with ApeKI and EcoT22I enzymes using the SimRAD package from R (Lepais & Weir, 2014). After selecting resultant digested fragments from 35 to 152 bp (to match the experimental data sizes), we recovered 35,462 and 32,827 sequences in S. purpuratus for ApeK1 and EcoT22I, respectively, and 160,087 and 17,038 in L. bergylta for ApeKI and EcoT22I respectively. The higher number of loci retrieved in the in silico digestion in comparison to the empirical data might be due to the large numbers of individuals genotyped in the population analyses (Carreras et al., 2020, 2021; Torrado et al., 2020). A reduction in the number of loci when increasing sample size has been previously reported due to the missing data filter (Casso et al., 2019). To infer the genomic composition of the obtained loci, we first mapped the selected digested sequences to their corresponding reference genome. As expected, the percentage of mapped sequences was higher than 99.9% in all cases (Table S3). To establish the genomic categories of the simulated loci, we only selected the sequences that mapped to unique positions to match the protocol followed with the categorization of the experimental dataset. We also estimated the frequency of the three categories (intergenic, intronic and exonic) in the genome. Most simulated loci mapped to genic regions for the two enzymes and species (Figure 3). The number of observed genic and intergenic loci for both enzymes and species was significantly different to those expected considering the genome composition (Table S4). Additionally, the abundance of genic regions observed in the genome was significantly different for digestions with ApeKI than with EcoT22I being higher in the former for both S. purpuratus (X2 = 284.9, p < .001) and L. bergylta (X2 = 264.9, p < .001). Moreover, the number of simulated loci in intergenic, exonic and intronic regions (Table S3) varied significantly between restriction enzymes in both S. purpuratus (X2 = 10,102, p < .001) and L. bergylta (X2 = 4491.4, p < .001). In particular, those simulated digestions with the ApeKI enzyme were enriched in exons, while those with the EcoT22I enzyme were enriched with introns (Figure 3, Table S3).
3.3 Functional analyses of genome-wide loci in fish and sea urchins
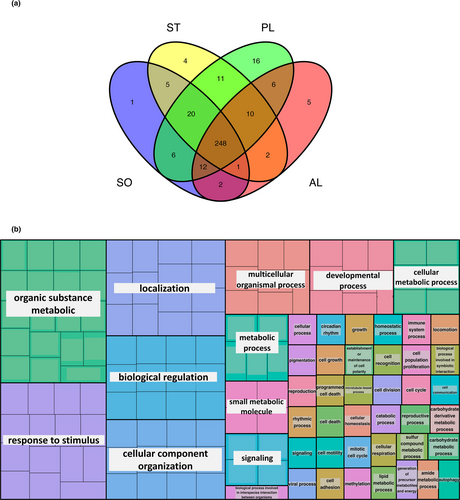
We performed functional analyses in order to characterize the loci that were mapped uniquely to genes in the corresponding reference genomes, by assigning GO terms to the longest isoform using eggNOG mapper software. The percentage of loci mapped uniquely to genes with assigned GO terms was 88.3%, 88.1%, 72.4% and 90.5% for S. ocellatus, S. tinca, P. lividus and A. lixula respectively. All species had a similar percentage in the root classification of GO terms, where the most abundant was “biological process” including between 77% and 80% of the GO terms (Figure S2). We classified the GO terms from the “biological process” category (3513, 3709, 4122, 2777 from S. ocellatus, S. tinca, P. lividus and A. lixula respectively) using the categories from GO slims (Table S5). Using GO slims, we were able to classify 99% of the GO terms obtained into 350 GO slims terms. The majority of the GO slims (70.8%) were shared between the four species (Figure 4a). The GO slims shared by the four species were involved in a myriad of basic mechanisms, such as response to stimulus, biological regulation, cellular component organization, etc. (Figure 4b and Table S5).
4 DISCUSSION
Genomics is revolutionizing our understanding of the adaptive capabilities of endangered species and aids management strategies by improving the delineation of conservation units (Funk et al., 2012). Candidate loci for adaptation, related to environmental cues, are often identified in population genomic studies after using a reduced representation sequencing technique (Benestan et al., 2016; Sandoval-Castillo et al., 2018; Torrado et al., 2022). The functional composition and gene category of candidate loci to be selected in several conditions and species have been studied in the past (Carreras et al., 2020; Pérez-Portela et al., 2020; Schunter et al., 2014; Torrado et al., 2022). However, the distribution of all analysed loci needed to be assessed in order to identify the processes leading to differences in genic distribution across studies and taxa. In the present work, we have shown that candidate loci obtained using the GBS technique are not enriched at certain genic categories but mirror the distribution of the total loci used in the population studies. By combining experimental and simulated datasets we determined that the genomic location of loci may be greatly influenced by the methodology used, especially in terms of the nucleotide content of the recognition sequence of the restriction enzyme. However, other factors, such as the divergence time to the reference genome, may play a role in the identification of loci at different genomic categories.
By mapping all loci to their closest available reference genome, we observed that most loci were located in genic regions in both experimental and simulated datasets. In the experimental datasets, we detected significant differences when comparing the proportion of loci mapping to exons and introns between groups. Sea urchin loci mostly mapped to exons, while fish loci mostly mapped to introns. This could be attributed to the different genome architecture of the two taxonomic groups (Galià-Camps et al., 2023). It is worth noting that the reduced representation technique used was the same for the four species (GBS), but the restriction enzyme used differed between groups: EcoT22I for fish, and ApeKI for sea urchins. In the simulated sets, where the two enzymes were assayed, we detected that the proportion of genic sites was significantly higher with ApeKI than EcoT22I and that the proportion of exonic regions was significantly enriched when cutting with ApeKI while the proportion of intronic regions was significantly enriched when cutting with EcoT22I. The restriction site of EcoT22I is (A | TGCA | T), thus the GC content of the target is only 33%. Conversely, the restriction site of ApeKI is (G | CWG | C), and the GC content represents 80% of the target. Knowing that exons have a higher percentage of GC content compared to introns (Amit et al., 2012; Kalari et al., 2006), it is expected that the loci obtained with the ApeKI enzyme (GC-rich) target a higher proportion of exonic regions, while the EcoTT22I enzyme (AT-rich) targets more non-exonic regions, such as introns and intergenic regions. Moreover, when comparing the genomic composition of the total and candidate loci within species, we did not detect any significant difference for any of the four species analysed, indicating that the candidate loci's composition mirrors the total loci distribution. Further studies are needed to confirm this result since in the empirical dataset the number of mapped loci was low due to the large phylogenetic distance to the closest reference genome. However, our simulated datasets are quite compelling indicating that the differential enrichment towards intronic and exonic regions detected in fish and sea urchins, respectively, seems to be due to the enzyme used for reduced representation sequencing of the genome and related to the GC content of the restriction site (Galià-Camps et al., 2023).
Previous studies (DaCosta & Sorenson, 2014; Kirschner et al., 2016; Roszik et al., 2017) also reported a bias caused by the restriction enzymes, especially towards first exons. Thus, the assumption of sequencing random fractions of the genome is not met, and it depends on the restriction enzyme selected. It is important to consider this finding when designing a study for conservation purposes. For instance, conservation studies focusing on adaptation in coding regions may benefit from GC-rich enzymes such as ApeKI, MspI, PstI, SbfI or SphI, while those focusing on neutral variability should select non-rich GC enzymes such as EcoT22I, EcoRI or MseI (see https://international.neb.com/tools-and-resources/selection-charts/isoschizomers for a broad list of restriction enzymes and their cut sites). However, it has been proposed that neutral and adaptive markers, which provide different types of evolutionary information, should be integrated to make optimal management decisions to protect biodiversity (Funk et al., 2012). Importantly, reduced representation sequencing, regardless of the enzyme used, provides clues on neutral and candidate adaptive markers by identifying outlier regions that help differentiate populations, either by finding the targets of selection or by linkage with selective loci (Carreras et al., 2017).
One of the striking results of our study is the low percentage of loci mapped to the closest available reference genome (less than 10%), likely a consequence of the divergence time between the reference genome species and the studied species. For instance, the percentage of loci mapped to the reference genome was higher in P. lividus than A. lixula (10% and 3% respectively), which is in agreement with their divergence time from S. purpuratus (58 MYA and 208 MYA, respectively, Figures 1 and S1). The number of microsatellites that successfully amplified in fish, negatively correlates with the phylogenetic distance to the source species (Carreras-Carbonell et al., 2008). Similarly, the number of reads mapping to a reference genome decreases according to the phylogenetic distance (Galla et al., 2018). Not only this, since genic regions are more conserved than intergenic regions of the genome (Chaffey, 2003), the more phylogenetically distant the focal and the source species of the reference genome, the more likely to target genic regions, as we observed in the present study. Thus, the use of phylogenetically distant reference genomes plus the usage of GC-rich restriction enzymes will increase the bias towards obtaining mapped loci in highly conserved genic regions, as we show in sea urchins. Finally, the quality of the genome, not only the assembly but also the annotation completeness are key when identifying loci. Despite the bias in genome composition, the functional analysis showed that most of the functions assigned to the mapped loci were shared between the four species analysed (Figure 4 and Table S5). Unfortunately, we could not perform a functional analysis of the candidate loci, due to the low percentage of loci mapped coupled with the lack of annotated GO terms in the reference genomes (annotations were transferred using orthology relationships). Altogether, conservation genomic studies based on reduced representation sequencing techniques will benefit from future high-quality and well-annotated reference genomes (Brandies et al., 2019; Formenti et al., 2022). Luckily, their availability is increasing due to several initiatives such as the ERGA consortium or the Earth Biogenome Project (Formenti et al., 2022). In addition, with the ever-increasing availability of public genomic datasets, in the future this study could be extended to a meta-analysis including other species, enzymes and reduced representation sequencing techniques.
5 CONCLUDING REMARKS
This study demonstrates that the selection of the restriction enzyme is key when using reduced representation sequencing techniques in conservation genomics studies. We obtained compelling evidence that restriction enzymes produce important differences in the composition of mapped loci. The analysis of simulated and experimental datasets obtained using two different restriction enzymes suggest that loci are biased towards exonic or intronic regions depending on the enzyme used. Although loci obtained are involved in a myriad of general functions, their functional composition seems to be affected by the loci targeted. The genome composition of candidate loci for adaptation mirrors one of the total loci in the four species analysed. Importantly, we show that the number of loci mapped and their characterization depends on the divergence time between the reference genome and the focal species, as well as, the reference genome quality. Our study highlights it is critical to select the restriction enzyme according to the biological question that aims to be addressed. In addition, the need for well-annotated reference genomes for non-model species to dig deep into the functionality of the candidate loci identified in population genomic studies aiming at species conservation.
AUTHOR CONTRIBUTIONS
All authors designed the research, analysed the data and contributed to writing the paper.
ACKNOWLEDGEMENTS
This research was funded by MarGeCh (PID2020-118550RB, funded by MCIN/AEI/10.13039/501100011033) from the Spanish Government. The authors CC, MP and CP are members of the research group SGR2021-01271 funded by the Generalitat de Catalunya.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
Genetic data were obtained from public repositories (A. lixula: PRJNA746276, P. lividus: PRJNA608661, Symphodus ocellatus: PRJNA646056 and Symphodus tinca: PRJNA646057). All the bioinformatic pipelines used in this research are available on GitHub (https://github.com/EvolutionaryGenetics-UB-CEAB/restrictionEnzimes.git).




