Control of Pseudomonas fluorescens in milk by a novel bacteriophage YZU_PF006
Abstract
Pseudomonas fluorescens is a psychrophilic bacterium that can cause dairy spoilage through the production of heat-stable enzymes. Bacteriophages are proved as one of the alternatives to control spoilage bacteria in the dairy industry. In this work, a lytic bacteriophage YZU_PF006 against P. fluorescens was isolated from sewage samples in China. Morphology and one-step growth results indicated that this short-tailed phage (a tail length of 5 nm and a head diameter of 58 nm) had a high burst size (180 PFU/infected cells) but a relatively short latent period (60 min). This phage can be inactive after pasteurisation and Clean-In-Place (CIP) procedures. The genomic sequence showed that phage YZU_PF006 had 47 open reading frames (ORFs) but did not have genes for virulence, lysogeny and antibiotic resistance. Phage YZU_PF006 effectively controlled P. fluorescens growth at 7°C and 28°C in milk, which indicates potential for the control P. fluorescens in the dairy industry.
INTRODUCTION
Pseudomonas fluorescens, a common psychrophilic and Gram-negative bacterium, is widely present in soil, air, water and other dairy farm environments. It can be easily introduced into raw milk, and extensive studies have consistently confirmed it as one of the dominant microorganisms in raw milk (Yuan et al. 2017; Du et al. 2022; Li et al. 2023; Yan et al. 2023a). This bacterium is also known as a primary contributor to dairy spoilage and waste due to the production of extracellular spoilage enzymes including proteases, lipases and β-galactosidases (Yuan et al. 2018). These spoilage enzymes are usually heat-resistant, and may negatively influence the quality of dairy products by causing colour defects, undesirable odours, bitterness, milk coagulation and reduction of yield in cheese manufacturing (Decimo et al. 2018; Zhang et al. 2019). Moreover, many works have consistently proved that P. fluorescens strains can quickly form thick biofilms on various food contact surfaces (Rossi et al. 2018; Yan et al. 2023a; Yuan et al. 2024). As an additional survival mechanism, the formed biofilm cells are difficult to be removed by Clean-In-Place (CIP) procedures, and usually act as a continuous bacterial contamination source to threaten the safety and sensory quality of commercial dairy products (Yuan et al. 2020, 2023).
Good hygienic practices at the farm level can decrease the microbial counts, but are not efficient to prevent the contamination of P. fluorescens in raw milk samples. Antibiotics are widely used for treating and preventing bacterial infections; however, the misuse of antibiotics promoted the spread of antibiotic-resistant Pseudomonas strains (Arslan et al. 2011). Therefore, there is an increasing need for natural antimicrobial substances as substitutes for chemical agents and antibiotics. Lytic phage interventions have been regarded as antibacterial agents against pathogenic and spoilage bacteria owing to the emergence of multidrug-resistant bacteria. Phages are viruses that can infect their bacteria hosts, multiply quickly and eventually lead to cell lysis with the release of the progeny (Mgomi et al. 2022). Phages are widespread in nature, and usually do not pose health risks to animals and humans, and do not affect the quality of foods. Therefore, phages have been widely considered as promising biocontrol agents to regulate the composition of their host bacterial communities in dairy environments (Ge et al. 2022; Zhang et al. 2022), for example, to eliminate foodborne pathogens distributed in various types of dairy products (García et al. 2009; Zhu et al. 2022; Zhang et al. 2023). In addition, some studies also confirmed the potential application of controlling the spoilage bacterium P. fluorescens in milk products (Yan et al. 2023a; Qin et al. 2024). However, due to P. fluorescens wide phenotypic and genotypic diversity in nature, the knowledge of the characterisation and application of phages to inhibit the growth of P. fluorescens is limited.
To address this knowledge gap, this work aimed to isolate a lytic phage against dairy derived P. fluorescens, and to determine its morphological feature, and biological features, including host range, pH and thermal stability, and one-step growth curve. The genomic property of phage YZU_PF006 was identified by genome sequencing.
MATERIALS AND METHODS
Bacteria strains and growth conditions
The host P. fluorescens P.f.4 and the other six P. fluorescens strains were previously isolated from raw milk samples, and stored in tryptic soy broth (TSB, Difco, Tucker, GA, USA) with 20% glycerol at –80°C. This bacterium was plated onto tryptic soy agar (TSA) plates, and then incubated at 28°C for 48 h. Thirty-two additional strains belonging to eight genera (Table 1) were selected for the host range of the newly isolated phage in this work. The above bacterial strains were grown in TSB or on TSA plates under optimum conditions. All the strains were provided by the Laboratory of Food Quality and Safety of Yangzhou University.
| Strains | Lytic |
|---|---|
| Pseudomonas azotoformans | − |
| Pseudomonas brenneri | − |
| Pseudomonas chlororaphis P.c.1 | − |
| Pseudomonas chlororaphis P.c.2 | − |
| Pseudomonas extreme orientalis | − |
| Pseudomonas fluorescens P.f.1 | + |
| Pseudomonas fluorescens P.f.3 | + |
| Pseudomonas fluorescens P.f.4 | + |
| Pseudomonas fluorescens P.f.5 | + |
| Pseudomonas fluorescens P.f.6 | + |
| Pseudomonas fluorescens P.f.7 | + |
| Pseudomonas fluorescens P.f.8 | + |
| Pseudomonas fulva | − |
| Pseudomonas gessardii P.g.1 | − |
| Pseudomonas gessardii P.g.2 | − |
| Pseudomonas gessardii P.g.3 | − |
| Pseudomonas gessardii P.g.4 | − |
| Pseudomonas gessardii P.g.5 | − |
| Pseudomonas grimontii P.g.t.1 | − |
| Pseudomonas grimontii P.g.t.2 | − |
| Pseudomonas koreensis P.k.1 | − |
| Pseudomonas koreensis P.k.2 | − |
| Pseudomonas koreensis P.k.3 | − |
| Pseudomonas libanensis | − |
| Pseudomonas mandelii | − |
| Pseudomonas marginalis | − |
| Salmonella typhimurium | − |
| Bacillus licheniformis | − |
| Enterobacter hormaechei | − |
| Escherichia coli | − |
| Staphylococcus aureus | − |
| Brochothrix thermosphacta | − |
| Listeria monocytogenes | − |
Phage YZU_PF006 isolation and purification
Domestic sewage sampled from Yangzhou was used for the isolation of P. fluorescens phages by the method provided by Chen et al. (2021) with slight modifications. Ten millilitres of each sewage sample was centrifuged at 8000 g for 10 min to remove any impurity, and the obtained supernatant was mixed with 100 μL of the culture of the host P. fluorescens P.f.4, and cultivated at 28°C for 24 h. Then, the culture was centrifuged at 4°C and 12 000 g for 10 min, and the obtained supernatant was filtered through 0.22 μm-filters. The double-overlay agar was applied for the screening of phage presence in each filtrate. Briefly, 100 μL of each filtrate was mixed with 100 μL host P. fluorescens P.f.4 at 108 cfu/mL in TSB soft agar, followed by incubation for 24 h at 28°C. After purification for at least three successive times, the individual transparent plaque was picked up, and transferred into SM buffer (100 mmol/L NaCl, 8 mmol/L MgSO4·7H2O, 50 mmol/L Tris–HCl), and mixed with 100 μL host P. fluorescens P.f.4 culture to form a double-layered plate. Phage titre was measured after incubation at 28°C for 24 h, and shown as plaque-forming units per millilitre (PFU/mL).
Morphology of phage YZU_PF006
Morphology of phage YZU_PF006 was observed by TEM by the method provided by Tang et al. (2023). Aliquots of 10 μL amplified phage suspension were deposited on a copper grid, absorbed for 10 min, stained with 2% phosphotungstic acid (Beijing Biotechnology Co., Ltd, Fuzhou, China) for 10 min, and the morphology of phage YZU_PF006 was obtained by TEM analysis (Tecnai).
Host range of phage YZU_PF006
The host range of phage YZU_PF006 was studied according to the spot testing assay (Zhang et al. 2023). In brief, 10 μL phage suspension was spotted onto each bacterial lawn, cultured for 24 h at 30°C, and an examined transparent plaque on plates indicated the host cell lysis.
Temperature and pH sensitivity of phage YZU_PF006
To explore the pH sensitivity of phage YZU_PF006, 100 μL phage suspensions (108 PFU/mL) were transferred to 900 μL of TSB with pH values ranging from 2 to 13 adjusted by NaOH or HCl, followed by the incubation for 2 h at 28°C (Chen et al. 2021). For the temperature sensitivity assay, 100 μL of each phage suspension (108 PFU/mL) were incubated at 40°C, 50°C and 60°C for 1 h, and then sampled at regular time intervals of 20 min (Tang et al. 2023). The titre of phage after each treatment was obtained according to the double-layer plate method.
One-step growth curve for phage YZU_PF006
The one-step growth curve for YZU_PF006 was performed according to the method by Tang et al. (2023) with modifications. The purified phage YZU_PF006 suspension (108 PFU/mL) and its host P. fluorescens P.f.4 culture (107 cfu/mL) was mixed at a MOI of 10 in TSB. After adsorption at 28°C for 10 min, the culture was centrifuged at 8000 g and 4°C for 10 min, and pellets were transferred into fresh TSB, and then incubated for 5 h at 28°C. Afterwards, 100 μL of aliquots were taken out at regular time intervals of 30 min, and the phage titre was measured by the double-layer agar plate method. The latent period, burst size and burst time were obtained from the one-step growth curve for phage YZU_PF006 by using DMFit web edition (http://modelling.combase.cc/DMFit.aspx) (Pujato et al. 2015).
Genome sequencing of phage YZU_PF006
Genome sequencing of phage YZU_PF006 was conducted by a method from Tang et al. (2023). In brief, the genomic DNA of phage YZU_PF006 was extracted and purified by the ABigen Lambda Phage Genomic DNA Kit (AB1141; ABigen Corporation, Beijing, China). The whole genome of phage YZU_PF006 was sequenced by the Illumina NovaSeq 6000 platform (Illumina, San Diego, CA, USA). Gene prediction and annotation were performed by GeneMarkS (http://topaz.gatech.edu/), and the National Center for Biotechnology Information database (NCBI, https://www.ncbi.nlm.nih.gov/). The phylogenetic tree of phage YZU_PF006 and other phages (Table S1) was constructed using MEGA X after sequence alignment, and modified by iTOL software (https://itol.embl.de/) (Chen et al. 2021).
Effect of phage YZU_PF006 against P. fluorescens in milk
In this study, the effect of phage YZU_PF006 against P. fluorescens in UHT milk was measured at both room temperature (28°C) and refrigerated temperature (7°C). In brief, 100 μL of the host P. fluorescens P.f.4 culture was inoculated into 10 mL of UHT milk to obtain a bacterial concentration of 104 cfu/mL. Afterwards, 100 μL phage suspensions with different concentrations (103 to 106 PFU/mL) were mixed with the cultures, and then incubated at 28°C for 24 h and 7°C for 192 h. After incubation, the cell count of P. fluorescens in each milk sample was measured by the plate counting method (Yan et al. 2023b).
Statistical analysis
In this study, every experiment was performed three times, and the statistical analysis was conducted by one-way analysis of variance (ANOVA) in Statistical Package for Social Sciences (SPSS version 20; IBM, Armonk, NY, USA). The statistical significance was considered when P < 0.05.
RESULTS AND DISCUSSION
Phage YZU_PF006 isolation and its morphology
Phages have been increasingly considered as promising alternatives to control pathogens and spoilage bacteria in different milk products (Zhu et al. 2022; Zhang et al. 2023). In previous studies, some P. fluorescens phages have been isolated and further used to control dairy spoilage caused by P. fluorescens (do Nascimento et al. 2022; Tayyarcan and Boyaci 2023; Yan et al. 2023b; Qin et al. 2024). However, due to its wide phenotypic and genotypic diversity in nature, the knowledge of the characterisation, and application of the phages to inhibit P. fluorescens growth and reduce dairy spoilage is not enough.
In the present work, the phage YZU_PF006 was isolated in sewage collected from Yangzhou. Upon the lawn of P. fluorescens P.f.4, phage YZU_PF006 formed clean plaques with a diameter of 0.5 cm (Figure 1a). According to the TEM micrograph (Figure 1b), phage YZU_PF006 was assigned to a short-tailed phage, since it showed a short tail with 5 nm length and a polyhedral head with a diameter of 58 nm. Similar to the previous results, most identified P. fluorescens phages were classified as a short-tailed phage (previously known as Podoviridae) (Qin et al. 2017; do Nascimento et al. 2022; Yan et al. 2023b).

Host range of phage YZU_PF006
In this work, a total of 33 strains from eight genera were selected to determine the host range of phage YZU_PF006 (Table 1). The results demonstrated that this phage YZU_PF006 only formed clear plaques on seven isolates of P. fluorescens, but did not exhibit its lytic ability against other species of Pseudomonas and other tested genera, which suggests it is highly specific for P. fluorescens strains. The reason for its inability to lyse other tested strains could be due to the specific binding of the tail filament to the receptor protein on the bacterial surface. Similarly, all P. fluorescens phages isolated in a previous work were proved to specifically infect the P. fluorescens host or a few Pseudomonas isolates (do Nascimento et al. 2022). Another study also showed that the P. fluorescens phage PD1 only showed the lytic activity against its host, but did not form plaques on other Pseudomonas and other genera isolates (Yan et al. 2023b). Although phages used as biocontrol agents are preferred to infect a large number of strains of the Pseudomonas genus, the preparation of phage cocktails can serve as an alternative way for phages with narrower host ranges.
Thermal and pH sensitivity of phage YZU_PF006
Different temperatures and pH values of the environment may significantly affect the viability and stability of phages, and their practical applications in foods or food processing environments (Chen et al. 2021; do Nascimento et al. 2022). In this study, both thermal and pH sensitivity of phage YZU_PF006 were studied according to the phage titres obtained from different conditions. The phage YZU_PF006 was relatively stable when tested at 40°C and 50°C for 60 min, and its titre only decreased by 0.14 log PFU/mL (40°C for 60 min) and 0.32 log PFU/mL (50°C for 60 min). However, its titre dramatically decreased after treatment at 60°C, and phage YZU_PF006 was totally inactive at 60°C for 20 min (Figure 2a). Previous studies also confirmed that most P. fluorescens are not heat-stable, and can be inactivated at 60°C (Qin et al. 2017; Tanaka et al. 2018). The above results suggested that this phage could be inactivated by pasteurisation after use, which also confirmed a reduced concern from consumers for the potential health risk caused by residual activity of phages in foods.
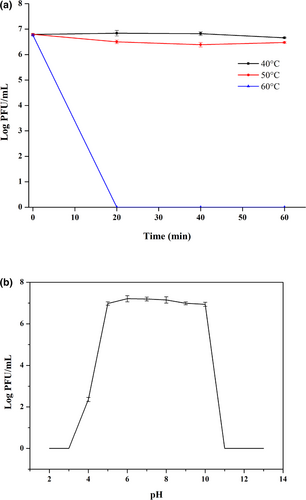
Regarding the pH stability, phage YZU_PF006 presented strong tolerance to pH from 5 to 10, but was decreased to 2.36 log PFU/mL at pH value of 4, and its activity was totally inactivated when pH values reached 1 and 11 (Figure 2b). The pH values at too high or too low levels usually affect the process of phage attachment to the receptor sited on the host cell. The pH stability of phage YZU_PF006 is lower when compared to the P. fluorescens phage PD1 isolated from fish guts, which remained highly active in buffers of pH 4–11 (Yan et al. 2023b). In another study, three P. fluorescens phages (L23.2, P22.1 and A11.1) were stable under neutral and alkaline conditions, but the phage titres were significantly reduced when the pH drops below 5 (Tayyarcan and Boyaci 2023). In general, the phages are usually exposed to the dairy environment with pH values from 4.5 to 6.8 (do Nascimento et al. 2022). The high stability of phage YZU_PF006 in a broad spectrum of pH values confirms potential application in the biocontrol of P. fluorescens in the dairy industry.
One-step growth curve for YZU_PF006
Application of phages for biocontrol is usually dependent on their growth characteristics including burst size, latent period and burst time. The one-step growth curve for phage YZU_PF006 clearly showed its latent period, rising period and plateauing period were 60, 30 and 180 min, respectively, and its burst size was 180 PFU/infected cells. The burst size of phage YZU_PF006 is rather higher, while its latent period is shorter than those of many reported P. fluorescens phages (Qin et al. 2017; Tanaka et al. 2018; do Nascimento et al. 2022; Yan et al. 2023b). For example, the latency period of P. fluorescens phage UFJF_PfDIW6 was 115 min, and its burst size was 145 PFU/infected cells (do Nascimento et al. 2022). The above results indicate the potential application of phage YZU_PF006 as a biocontrol agent, since it may quickly replicate, and effectively inactive its target bacteria.
Genome characterisation of the phage YZU_PF006
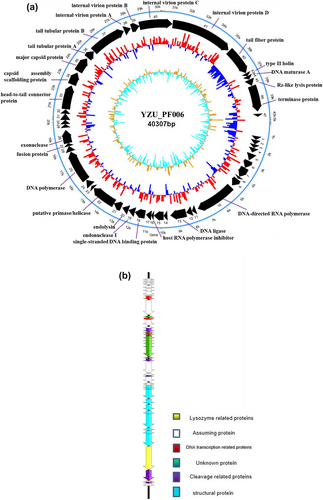
Genomic characterisation can provide molecular traits of phages, and help for designing effective biocontrol agents against phage. The genomic features of YZU_PF006 are shown in Figure 3(a). The genome of phage YZU_PF006 is double-stranded with a total length of 40 307 bp, and an average G + C content of 59.93%. Genome sizes of P. fluorescens phages in other studies showed their genome sizes ranged from 38 758 bp to 42 478 bp, and with a G + C content ranged from 21.16 to 58.3% (Hungaro et al. 2022; Vidigal and Hungaro 2023; Yan et al. 2023b). From ORF1 to ORF47 in its genome, 40 ORFs were functionally predicted according to homology proteins available in the database, among which, 24 had annotated functions and 17 were annotated as hypothetical proteins (Table 2). The completed phage genome sequence of phage YZU_PF006 was deposited in GenBank (accession number PP704628).
| ORFs | Putative function | Best phage homologue (identities %) |
|---|---|---|
| 10 | DNA-directed RNA polymerase | Pseudomonas virus WRT (98.8%) |
| 13 | DNA ligase | Pseudomonas phage phiPsa17 (98.3%) |
| 16 | Host RNA polymerase inhibitor | Pseudomonas phage phiPsa17 (98.2%) |
| 18 | Single-stranded DNA binding protein | Pseudomonas virus Pf1 ERZ-2017 (95.3%) |
| 19 | Endonuclease I | Pseudomonas phage phiPSA2 (100%) |
| 20 | Endolysin | Pseudomonas phage phiPSA2 (97.9%) |
| 22 | Putative primase/helicase | Pseudomonas phage phiPSA2 (96.3%) |
| 25 | DNA polymerase | Pseudomonas virus KNP (99%) |
| 27 | Fusion protein | Pseudomonas phage phiPSA2 (97.1%) |
| 28 | Exonuclease | Pseudomonas virus Pf1 ERZ-2017 (98.6%) |
| 33 | Head-to-tail connector protein | Pseudomonas virus KNP (98.9%) |
| 34 | Capsid assembly scaffolding protein | Pseudomonas virus KNP (99%) |
| 35 | Major capsid protein | Pseudomonas phage phiPSA2 (100%) |
| 36 | Tail tubular protein A | Pseudomonas phage gh-1 (99%) |
| 37 | Tail tubular protein B | Pseudomonas virus WRT (96.4%) |
| 38 | Internal virion protein A | Pseudomonas phage gh-1 (96.5%) |
| 39 | Internal virion protein B | Pseudomonas phage phiPsa17 (96.4%) |
| 40 | Internal virion protein C | Pseudomonas phage phiPsa17 (96.5%) |
| 41 | Internal virion protein D | Pseudomonas phage gh-1 (97.8%) |
| 42 | Tail fibre protein | Pseudomonas virus WRT (75.2%) |
| 43 | Type II holin | Pseudomonas phage PPpW-4 (98.5%) |
| 44 | DNA maturase A | Pseudomonas phage shl2 (80%) |
| 45 | Rz-like lysis protein | Pseudomonas phage phiPSA2 (65.1%) |
| 46 | Terminase protein | Pseudomonas phage phiPSA2 (97.9%) |
Despite that about 50% of proteins had no predicted functions, the putative ORFs identified in phage YZU_PF006 are classified into four modules according to their predicted functions (Figure 3b). Ten ORFs are homologous genes related to phage structures and morphology, for example, ORF33 (head-to-tail connector protein), ORF35 (major capsid protein), ORF36 (tail tubular protein A), ORF37 (tail tubular protein B) and ORF42 (tail fibre protein). Capsid proteins in phages are important structures of the virion, and may shelter the viral genome from extreme environments. Phage tail structures are usually responsible for the recognition and penetration of hosts, and channel formation for phage genome injection. The two-tail tubular proteins (A and B) have also been identified in short-tailed phage genomes. Three ORFs, such as ORF20 (endolysin), ORF43 (type II holin) and ORF45 (Rz-like lysis protein), are predicted for the function of host lysis. Both endolysin and holing proteins were also found in P. fluorescens phage PD1 (Yan et al. 2023b). At the late stage of multiplication, phages usually release phage progeny from the host based on an ‘endolysin-holin’ lysis system (Zhang et al. 2023). In this work, holin (ORF43) can act on the cell membrane of the host, and then endolysin (ORF20) may reach the cell wall of bacteria. These three ORFs, in combined with the ORF42 (tail fibre protein), conferred the capacity of phage YZU_PF006 to specifically recognise and infect its host bacteria P. fluorescens P.f.4, which is in accord with the previous work (Yan et al. 2023b). Moreover, 7 and 3 ORFs are homologous genes annotated for the functions of DNA packaging and DNA metabolism, respectively. ORF19 (endonuclease I) and ORF28 (exonuclease) may hydrolyse DNA and provide deoxyribonucleotides for DNA synthesis of phages. Among the DNA packaging ORFs, the function of terminase protein (ORF46) is to recognise and cleave the viral DNA, and its translocation into a preformed empty phage capsid by ATP hydrolysis. Importantly, the genome of this phage YZU_PF006 does not contain any genes for virulence, lysogeny and antibiotic resistance. This is also an important feature for its potential application in the dairy industry. However, the other ORFs with unknown functions still have to be elucidated in the future. The above genomic features of phage YZU_PF006 suggested the high genetic diversity of P. fluorescens phages.
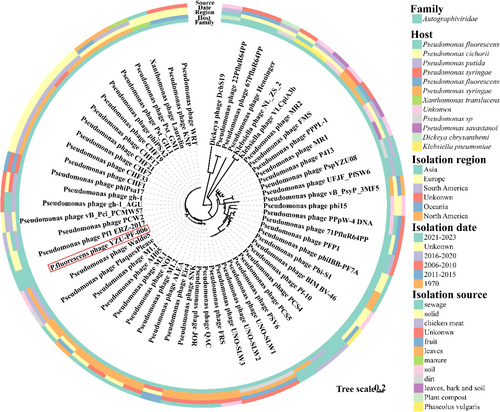
In addition, 55 P. fluorescens phages and four phages belonging to other genera with homology to phage YZU_PF006 were picked for the phylogenetic analysis of their genomes (Figure 4). The genomic DNA of phage YZU_PF006 presented a significant degree of sequence identity to Pseudomonas phage Pf1 ERZ-2017 (GenBank: NC_047874.1, highest homology: 94.28%) from sewage in Russia. The above genomic features of phage YZU_PF006 suggested the high genetic diversity of P. fluorescens phages.
Inhibition of P. Fluorescens in UHT milk by phage YZU_PF006
The effect of phage YZU_PF006 against P. fluorescens P.f.4 was tested in UHT milk at both 28 and 7°C (Figure 5). The total cell counts of P. fluorescens P.f.4 increased to 8.41 log cfu/mL in UHT milk without phage after incubation, but decreased (p < 0.05) to about 2 log cfu/mL after treatment by different phage concentrations (MOI at 0.01, 0.1, 1, 10 and 100) at 28°C for 24 h (Figure 5a). When treated at 7°C, the total cell counts of P. fluorescens P.f.4 increased to 8.02 log cfu/mL in UHT milk without phage after incubation at 7°C for 192 h (Figure 5b). The cell counts of P. fluorescens P.f.4 in UHT milk were decreased (p < 0.05) to the range of 2.38–4.34 log cfu/mL after phage treatment (MOI at 0.01, 0.1, 1, 10 and 100) at 48 h and 7°C, and this inhibitory was shown to be a phage concentration-dependent manner. However, the P. fluorescens cells were increased after incubation at 7°C for 48 h, and increased to 6.75–8.04 log cfu/mL at 192 h. Tayyarcan and Boyaci (2023) determined the effect of P. fluorescens phage on the evolution of phage resistance of hosts. It was observed that the lytic activity of the phage did not show a clear lytic activity against four hosts, which indicated that some of the hosts may develop a resistance (Tayyarcan and Boyaci 2023).
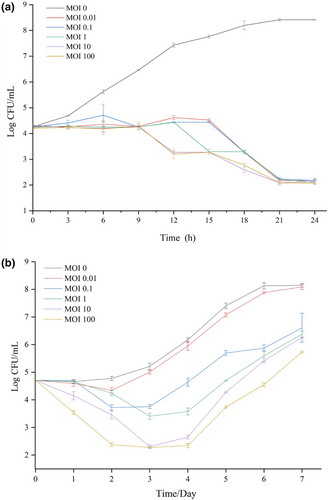
Results from previous reports have also confirmed that the strong lytic ability of P. fluorescens phages could inhibit Pseudomonas growth at low temperatures in milk. Yan et al. (2023b) reported that the cell counts of P. fluorescens in milk were reduced by up to 2.71 and 2.84 log cfu/mL when treated by its phage P. fluorescens PD1 with MOI of 100 and 1000 at 25°C. In another work, do Nascimento et al. (2022) confirmed that lytic phages of UFJF_PfDIW6 and UFJF_PfSW6 reduced bacterial count in milk at 4°C and 10°C, and inhibition of P. fluorescens proteases production for least 2 days.
CONCLUSION
The results of this work provide information on the morphological, biological and genomic characteristics of a novel phage YZU_PF006 that can infect P. fluorescens. Moreover, the strong inhibitory effect of phage YZU_PF006 against P. fluorescens in milk at refrigeration temperatures was also proved. In conclusion, this newly isolated phage YZU_PF006 could be used alone or in combination with other control strategies to serve as one of the good antimicrobial candidates to control the P. fluorescens contamination in dairy processing environments, and to promote both the safety and sensory quality of raw milk and milk products. In the future, more promising phage isolations of P. fluorescens are still needed for the dairy industry.
ACKNOWLEDGEMENTS
This work was supported by grants from the Natural Science Foundation of Jiangsu Province (Grants No. BK20210814), the National Natural Science Foundation of China (Grants No. 32302953), the Natural Science Fund for Colleges and Universities in Jiangsu Province (Grants No. 21KJB550007), China Postdoctoral Science Foundation (2022M720120; 2021TQ0274) and ‘QingLan’ Talent Support Program of Yangzhou University.
AUTHOR CONTRIBUTIONS
Lei Yuan: Investigation; methodology; writing – original draft; funding acquisition. Xinhai Yuan: Investigation; methodology; validation. Caowei Chen: Methodology; data curation. Wenyuan Zhou: Methodology. Zhenbo Xu: Methodology; formal analysis. Thanapop Soteyome: Data curation; writing – review and editing. Zhenquan Yang: Supervision; writing – review and editing.
CONFLICT OF INTEREST
All authors have no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
All data that support the results of this work are available from the corresponding author upon reasonable request.




