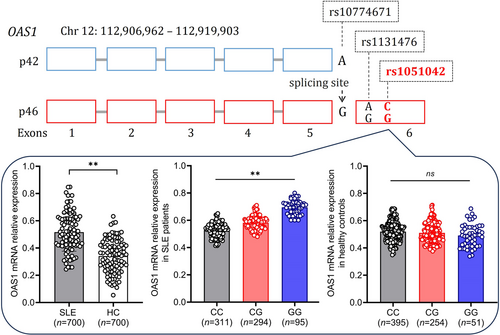
Association of OAS1 rs1051042 polymorphism with genetic susceptibility and clinical phenotypes to systemic lupus erythematosus in a Chinese population
Edited by Zhiyu Wang and Lishao Guo
Graphical Abstract
OAS1 is an interferon-induced gene that has been reported to be associated with the susceptibility of some infectious diseases, but there is little data on OAS1 single nucleotide polymorphism (SNP) associated with Systemic lupus erythematosus (SLE). In this study, we evaluated the association of OAS1 rs1051042 polymorphism with the genetic susceptibility and clinical phenotypes of SLE. Our results revealed patients with CG and GG genotypes were more susceptible to SLE. The genotype CG, GG, and allele G were associated with increased genetic susceptibility of SLE. OAS1 rs1051042 polymorphism was associated with clinical manifestations of SLE including malar rash, lupus nephritis, and anti-dsDNA antibodies. The expression levels of OAS1 were more significantly increased in genotype GG than in genotype CC and CG. Our results indicate polymorphism of OAS1 rs1051042 was associated with the genetic susceptibility and specific clinical phenotypes of SLE in the Chinese population.
Summary
-
Polymorphism of OAS1 rs1051042 is associated with the genetic susceptibility and clinical phenotypes of Systemic Lupus Erythematosus (SLE) in the Chinese population.
-
The genotype CG/GG and allele G of SNP rs1051042 in OAS1 are associated with increased genetic susceptibility of SLE.
-
OAS1 rs1051042 polymorphism is associated with specific clinical phenotypes of SLE including malar rash, lupus nephritis, and anti-dsDNA antibodies.
-
The expression levels of OAS1 are more significantly increased in genotype GG than in genotype CC and CG of SNP rs1051042.
Systemic lupus erythematosus (SLE) is a chronic autoimmune disorder characterized by multiple organs and systems damage.1, 2 Single-nucleotide polymorphism (SNP) studies have identified a host of loci associated with susceptibility and clinical manifestations of SLE.3-5 Oligoadenylate synthetases (OAS) are one family of interferon-induced genes and are activated in the presence of double-stranded or single-stranded RNA with secondary structures and catalyzes the synthesis of 2′,5′ linked oligoadenylate. OAS1 SNP rs1051042 is located in exon 6, and the earlier genetic studies showed that rs1051042 is a lead expression quantitative trait locus, suggesting potential causality of the OAS1 gene. However, there is no study on the association of OAS1 rs1051042 polymorphism and genetic susceptibility to SLE and other autoimmune diseases. In this study, we typed this SNP among our SLE cohort and aimed to investigate the potential association between the OAS1 rs1051042 polymorphism and SLE susceptibility in a Chinese population. In addition, we also analyzed the association of OAS1 rs1051042 polymorphism with the expression levels of OAS1 in SLE patients.
A total of 700 SLE patients were recruited from The Second Xiangya Hospital of Central South University between January 1, 2010 to January 1, 2024. All patients were diagnosed according to the 2012 Systemic Lupus Erythematosus International Collaborating Clinics (SLICC) classification criteria for SLE. All healthy controls had no history of autoimmune diseases and serious infections. Genotyping of SNP was performed by TaqMan SNP Genotyping Assay Kit (ThermoFisher) according to the manufacturer's instructions. The expression levels of OAS1 mRNA were detected by quantitative real-time polymerase chain reaction (PCR) using SYBR Premix Ex TaqTM (TaKaRa) and Roche-LightCycler96 Real-Time PCR System (Basel).
Alleles and genotypes were compared between different study groups using the chi-square test or Fisher's exact test. Estimated odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated using a 2 × 2 contingency table. One-way analysis of variance was used to analyze the data from OAS1 expression. The data were analyzed by SPSS 23.0 software. Significant differences were considered when the p-value was lower than 0.05.
Baseline characteristics of SLE patients and healthy controls in our cohort are shown in Supporting Information: Table S1. The sex ratio between the two groups (625 females and 75 males in patient group vs. 610 females and 90 males in control group) showed no significant difference (χ2 = 0.113, p = 0.737). The median age between SLE patient group and healthy control group (34.22 ± 3.22 years vs. 34.13 ± 7.14 years) was also no significant difference (t = 0.304, p = 0.761). The clinical phenotypes of SLE patients in this cohort were shown in Supporting Information: Table S1. The median score of the SLE disease activity index was 17.88 ± 2.11, and the median age of disease onset was 19.24 ± 4.34 years. The most common clinical manifestation was lupus nephritis (n = 611, 87.29%). The most common autoantibodies were antinuclear antibody (n = 700, 100.00%) and anti-Smith (n = 619, 88.43%).
The genotype CC (n = 311, 44.43%), CG (n = 294, 42.00%), and GG (n = 95, 13.57%) in SLE patients appeared to have a significantly different frequency as compared to CC (n = 395, 56.43%), CG (n = 254, 36.29%) and GG (n = 51, 7.29%) in healthy controls (χ2 = 26.174, p < 0.001) (Table 1). The genotype CG (CG vs. CC: OR = 1.470; 95% CI: 1.175–1.838, p = 0.001) and GG (GG vs. CC: OR = 2.366; 95% CI: 1.632–3.429, p < 0.001) were associated with significantly increased susceptibility of SLE when compared to the genotype CC. The allele C (916, 54.43%) and G (484, 34.57%) in SLE patients appeared to have a significantly different frequency as compared to C (1044, 74.57%) and G (356, 25.43%) in healthy controls (χ2 = 27.864, p < 0.001). The allele G was associated with significantly increased susceptibility to SLE when compared to the allele C (G vs. C: OR = 1.550; 95% CI: 1.316–1.825, p < 0.001). In the dominant model, the genotype GG (GG vs. CC + CG: OR = 1.998; 95% CI: 1.397–2.858, p < 0.001) was associated with significantly increased susceptibility of SLE. In the recessive model, the genotype CG + GG (CG + GG vs. CC: OR = 1.620; 95% CI: 1.312–2.001, p < 0.001) was associated with significantly increased susceptibility of SLE. In the heterozygote model, the genotype GG (GG vs. CG: OR = 1.609; 95% CI: 1.101–2.352, p = 0.014) was associated with significantly increased susceptibility of SLE. In the homozygote model, the genotype GG (GG vs. CC: OR = 2.366; 95% CI: 1.632–3.429, p < 0.001) was associated with significantly increased susceptibility of SLE. Together, our results revealed that patients with genotype CG/GG or allele G were more susceptible to SLE.
| OAS1 rs1051042 | SLE group (n = 700) | Control group (n = 700) | χ2 | p-value | OR (95% CI) | p-value |
|---|---|---|---|---|---|---|
| Genotype | ||||||
| CC | 311 (44.43) | 395 (56.43) | 26.174 | <0.001 | Reference | |
| CG | 294 (42.00) | 254 (36.29) | 1.470 (1.175–1.838) | 0.001 | ||
| GG | 95 (13.57) | 51 (7.29) | 2.366 (1.632–3.429) | <0.001 | ||
| Allele | ||||||
| C | 916 (54.43) | 1044 (74.57) | 27.864 | <0.001 | Reference | |
| G | 484 (34.57) | 356 (25.43) | 1.550 (1.316–1.825) | <0.001 | ||
| Dominant model | ||||||
| CC + CG | 605 (86.43) | 649 (92.71) | 14.804 | <0.001 | Reference | |
| GG | 95 (13.57) | 51 (7.29) | 1.998 (1.397–2.858) | <0.001 | ||
| Recessive model | ||||||
| CC | 311 (44.43) | 395 (56.43) | 20.161 | <0.001 | Reference | |
| CG + GG | 389 (55.57) | 305(43.57) | 1.620 (1.312–2.001) | <0.001 | ||
| Heterozygote model | ||||||
| CG | 294 (42.00) | 254 (36.29) | 6.102 | 0.014 | reference | |
| GG | 95 (13.57) | 51 (7.29) | 1.609 (1.101–2.352) | 0.014 | ||
| Homozygote model | ||||||
| CC | 311 (44.43) | 395 (56.43) | 21.424 | <0.001 | reference | |
| GG | 95 (13.57) | 51 (7.29) | 2.366 (1.632–3.429) | <0.001 |
- Abbreviations: CI, confidence interval; OR, odds ratio; SLE, systemic lupus erythematosus.
We conducted stratification analysis by comparing allele and genotype frequencies between positive and negative patients in 17 specific clinical phenotypes of SLE patients in this cohort (Supporting Information: Table S2). The results showed the significant associations of OAS1 rs1051042 polymorphism with three specific clinical phenotypes of SLE patients including malar rash (χ2 = 9.579, p = 0.008), lupus nephritis (χ2 = 25.064, p < 0.001), and anti-dsDNA antibodies (χ2 = 134.431, p < 0.001). On the contrary, we failed to detect any marked associations of OAS1 rs1051042 polymorphism with other listed clinical phenotypes (All p > 0.05, Supporting Information: Table S2).
The expression levels of OAS1 mRNA in all three genotypes were significantly different in SLE patients and most significantly increased in genotype GG than in genotype CC and CG (p < 0.001). There was no difference between the three genotypes in the control group (p > 0.05) (Figure 1). These data indicated that rs1051042 polymorphism regulated OAS1 expression in SLE, and patients with genotype GG, as well as the highest levels of OAS1 may be more susceptible to SLE.
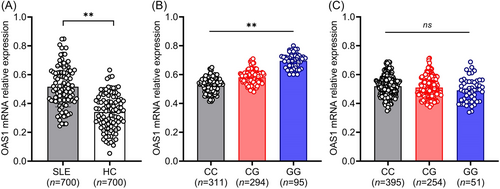
However, only one SNP was investigated, and selection bias from hospital-based revealed some limitations in this study. To elucidate the accurate mechanism of OAS1 variants in SLE, a larger sample size and principal component analysis are needed in the future. In conclusion, the OAS1 rs1051042 polymorphism showed an association with the genetic susceptibility and specific clinical phenotypes of SLE in the Chinese population. The analysis of genotype and allele frequency highlighted that genotype CG/GG of OAS1 rs1051042 was associated with higher expression of OAS1 and the susceptibility to malar rash, lupus nephritis, and anti-dsDNA antibodies. These results may help us identify a novel genetic factor for SLE patients and provide new insights into therapeutic strategies for SLE.
AUTHOR CONTRIBUTIONS
Xin Huang conducted the statistical analysis, RNA isolation, quantitative real-time PCR and drafted the manuscript. Ruifang Wu, Miao Yang, and Qianwen Li helped with the manuscript writing. Shuaihantian Luo designed the study, helped with statistical analyses, and revised the manuscript. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS
This study was supported by the Natural Science Foundation of Hunan Province, China, Grant/Award Numbers: 2021JJ40856, 2023JJ40827. The authors thank Prof. Qianjin Lu, Prof. Ming Zhao, Prof. Haijing Wu, and all students in the Hunan Key Laboratory of Medical Epigenomics for their efforts to collect blood samples.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
ETHICS STATEMENT
This study was approved by the Ethics Committee of the Second Xiangya Hospital of Central South University (Approval number: 2024-Z0622-01). Written informed consent was obtained from all subjects.
Open Research
DATA AVAILABILITY STATEMENT
All relevant data are within the manuscript and its additional files.