Crystal structure of an aspartate aminotransferase (TM1255) from Thermotoga maritima at 1.90 Å resolution
The TM1255 gene of Thermotoga maritima encodes an aspartate aminotransferase (AspC-1, EC 2.6.1.1), with a molecular weight of 42,287 Da (residues 1–377) and a calculated isoelectric point of 6.8. Aminotransferases contain a pyridoxal 5′-phosphate (PLP) cofactor and are involved in the degradation of most amino acids where they catalyze the transfer of their α-amino group to α-keto acids. Aspartate aminotransferase (AspAT), one of the most important of these enzymes, catalyzes a reversible transamination reaction between the dicarboxylic α-amino and α-keto acids by a ping-pong bi-bi mechanism.1, 2 AspATs from many species have been structurally characterized and classified into subgroups Ia and Ib.3-5 TM1255 and its homologue from Thermus thermophilus share 43% sequence identity and belong to subgroup Ib.3 Both share less than 18% sequence identity to the AspATs in subgroup Ia. According to sequence comparisons, most of the active-site residues essential for catalysis are conserved between AspATs of subgroups Ia and Ib, but the residue that recognizes the distal carboxylate of the substrate (Arg292 in AspATs of subgroup Ia) is replaced by Lys 101 in the AspATs of subgroup Ib.5 Here, we report the crystal structure of TM1255 with a covalently bound PLP cofactor molecule determined using the semiautomated high-throughput pipeline of the Joint Center for Structural Genomics (JCSG).6
The structure of TM1255 [Fig. 1(A)] was determined to 1.90 Å resolution with the molecular replacement (MR) method, using a search model constructed from AspAT from T. thermophilus, with a sequence identity of 43% [Protein Data Bank (PDB) code: 1BKG].5 Data collection, model, and refinement statistics are summarized in Table I. The final model includes 2 protein molecules (residues 2–376 for chain A, residues 1–376 plus 8 residues from the N-terminal His-tag for chain B), 2 covalently bound PLP molecules, 10 sulphates, and 641 water molecules. No electron density was observed for residue 377 in both chains. The Matthews' coefficient (Vm) for TM1255 is 2.31 Å3/Da, and the estimated solvent content is 46.3%. The Ramachandran plot, produced by PROCHECK 3.4,7 shows that 91% of the residues are in the most favored regions, and 9% are in additional allowed regions.
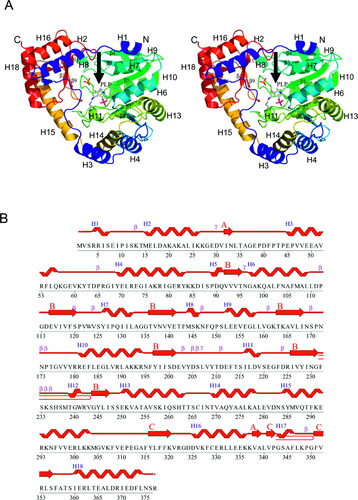
Crystal structure of TM1255. (A) Stereo ribbon diagram of Thermotoga maritima TM1255 color coded from N-terminus (blue) to C-terminus (red) showing the domain organization and the location of the active site (arrow). The helices H1–H18 and β-strands (β1–β12) are indicated. The pyridoxal 5′-phosphate (PLP) cofactor molecule covalently attached to Lys234 is depicted in ball and stick. (B) Diagram showing the secondary structure elements in TM1255 superimposed on its primary sequence. β-hairpins are depicted in red and β-sheets labelled A–C.
| Space group | P212121 | ||
| Unit cell parameters | a = 64.05 Å, b = 79.51 Å, c = 170.18 Å, α = β = γ = 90° | ||
| Data collection | λ0 | ||
| Wavelength (Å) | 0.900 | ||
| Resolution range (Å) | 43.00–1.90 | ||
| Number of observations | 249,027 | ||
| Number of reflections | 59,510 | ||
| Completeness (%) | 85.8 | ||
| (In highest resolution shell, %) | 50.6 | ||
| Mean I/σ (I) | 15.9 | ||
| (In highest resolution shell) | 1.7 | ||
| Rsym on I | 0.063 | ||
| (In highest resolution shell) | 0.422 | ||
| Sigma cutoff | 0.0 | ||
| Highest resolution shell (Å) | 2.00–1.90 | ||
| Model and refinement statistics | |||
| Resolution range (Å) | 43.00–1.90 | Data set used in refinement | λ0 |
| No. of reflections (total) | 59,510 | Cutoff criteria | |F| > 0 |
| No. of reflections (test) | 1249 | Rcryst | 0.159 |
| Completeness (% total) | 85.8 | Rfree | 0.200 |
| Stereochemical parameters | |||
| Restraints (RMS observed) | |||
| Bond length | 0.017 Å | ||
| Bond angle | 1.62° | ||
| Average isotropic B-value | 31.1 Å2 | ||
| ESU based on R value | 0.15 Å | ||
- ESU = Estimated overall coordinate error.14, 18
- Rsym = Σ|I1−〈I1〉|/Σ|Ii| where Ii is the scaled intensity of the ith measurement, and 〈Ii〉 is the mean intensity for that reflection.
- Rcryst = Σ||Fobs| − |Fcalc||/Σ|Fobs|, where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
- Rfree = as for Rcryst, but for 2.1% of the total reflections chosen at random and omitted from refinement.
The TM1255 monomer is composed of 12 β-strands (β1–β12) and 19 helices (H1–H19), where H2, H5, H9, and H14 are 310-helices [Fig. 1(A and B)]. The total β-strand, α-helical, and 310-helical content is 14.4%, 42.1%, and 2.9%, respectively. According to the Structural Classification of Proteins database (SCOP),8 TM1255 is a member of the “PLP-dependent transferase” fold. Like other AspATases TM1255 folds into the N-terminal arm (residues 1–11), the small domain (residues 12–40 and 283–376), and the large domain (residues 41–282) [Fig. 1(A and B)]. The small domain is an α/β-domain that contains 4 α-helices (H2, H15, H16, H18) and 2 antiparallel β-sheets, A and C, comprised of β-strands β1, β10 and β9, β11, β12, respectively. The large domain is an α/β/α-domain with a mixed β-sheet of 7 strands (β2–β8). The 7 β-strands show 2876534 topology and are all parallel except for β8. The β-sheet (B) is twisted and surrounded by 10 α-helices [Fig. 1(A and B)]. TM1255 forms a dimer through the interaction of its arm and the large domain [Fig. 2(A)]. The dimer interface corresponds to interactions between α-helices H1, H3–H5, and H10–12, with a buried surface area of 1958 Å2 per monomer. Each TM1255 monomer forms an active-site pocket between the small and the large domain [Figs. 1(A) and 2(B)]. The PLP cofactor resides at the bottom of the active-site pocket and forms a Schiff base with lysine 234. In addition, the active site contains a sulfate ion in close proximity to the cofactor [Fig. 2(C)].
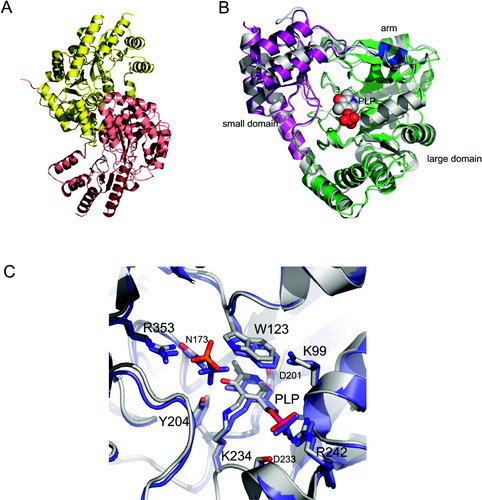
(A) Ribbon diagram of the TM1255 dimer. (B) Ribbon diagram of a superposition of TM1255 with its structural homologue AspAT from Thermus thermophilus (PDB code: 1BJW; white). TM1255 is shown with the arm, the small, and the large domain colored in blue, purple, and green, respectively. For a reference the pyridoxal 5′-phosphate (PLP) from TM1255 is shown in cpk mode. (C) Superposition of the active sites in TM1255 (white) and AspAT from T. thermophilus (blue). The active site residues, the PLP molecule, and the sulphates (phosphate in AspAT from T. thermophilus) are shown in ball and stick. The atoms for TM1255 are indicated as follows: carbon (gray), nitrogen (blue), oxygen (red), phosphor (purple), and sulfur (orange).
A structural similarity search, performed with the coordinates of TM1255 using the DALI server,9 indicates that the closest structural homologue is AspAT from T. thermophilus (PDB code: 1BJW).5 The root-mean-square deviation (RMSD) between TM1255 and AspAT from T. thermophilus is 1.3 Å over 370 aligned residues with 45% sequence identity. The main differences between TM1255 and AspAT from T. thermophilus are a 1.1-Å displacement of α-helix H2 and the α-helix H15 being one helical turn shorter in TM1255. All active-site residues essential for catalysis are conserved, which includes the counterpart of Lys101 (Lys99 in TM1255) that recognizes the side-chain carboxylate of the substrate in type Ib AspATs [Fig. 2(C)].
According to the Fold and Function Assignment System (FFAS),10 TM1255 has at least 15 distant homologues in the Thermotoga proteome. Six of them are annotated as aminotransferases: TM1698 (aspC-2) with 27% sequence identity, TM1040 (hisC) with 12% sequence identity, TM1131 with 22% sequence identity, TM1692 with 15% sequence identity, TM1400 with 15% sequence identity, and TM1371 (nifS) with 14% sequence identity. Models for TM1255 homologues can be accessed at http://www1.jcsg.org/cgi-bin/models/get_mor.pl?key=tm1255.
The structure reported here represents the second thermophilic AspAT with a covalently bound PLP cofactor, whose structure has been determined by X-ray crystallography. The information reported here, in combination with further biochemical and biophysical studies, will yield valuable insights into the functional determinants of this protein family and the thermostability of these organisms.
Materials and Methods.
Protein production and crystallization: TM1255 (AspC-1; TIGR: TM1255; Swissprot: Q9X0Y2) was amplified by PCR from T. maritima strain MSB8 genomic DNA using PfuTurbo (Stratagene) and primer pairs encoding the predicted 5′- and 3′-ends of TM1255. The polymerase chain reaction (PCR) product was cloned into plasmid pMH1, which encodes expression and purification tags consisting of the amino acids MGSDKIHHHHHH at the amino terminus of the full-length protein. The cloning junctions were confirmed by sequencing. Protein expression was performed in a modified Terrific Broth [24 g/L yeast extract, 12 g/L tryptone, 1% (v/v) glycerol, 50 mM 3-(N-morpholino) propanesulfonic acid (MOPS) pH 7.6] using the Escherichia coli methionine auxotrophic strain DL41. Lysozyme was added to the culture at the end of fermentation to a final concentration of 1 mg/mL. Bacteria were lysed by sonication after a freeze-thaw procedure in Lysis Buffer [50 mM Tris pH 7.9, 50 mM NaCl, 1 mM MgCl2, 0.25 mM Tris (2-carboxyethyl) phosphine hydrochloride (TCEP)], and the cell debris was pelleted by centrifugation at 3400 × g for 60 min. The soluble fraction was applied to a metal chelate affinity resin (Amersham Biosciences) previously charged with nickel and equilibrated with Equilibration Buffer [50 mM potassium phosphate pH 7.8, 0.25 mM TCEP, 10% (v/v) glycerol, 300 mM NaCl] containing 20 mM imidazole. The resin was washed with Wash Buffer [50 mM potassium phosphate pH 7.8, 300 mM NaCl, 40 mM imidazole, 10% (v/v) glycerol, 0.25 mM TCEP], and the protein was eluted with Elution Buffer [20 mM Tris pH 7.9, 300 mM imidazole, 10% (v/v) glycerol, 0.25 mM TCEP]. Buffer exchange was performed to remove imidazole from the eluate, and the protein in Buffer A [20 mM Tris pH 7.9, 5% (v/v) glycerol, 0.25 mM TCEP] containing 50 mM NaCl was applied to a Resource Q column (Amersham Biosciences) previously equilibrated with the same buffer. Protein was eluted using a linear gradient of 50–500 mM NaCl in Buffer A, and appropriate fractions were pooled. Protein was buffer exchanged into crystal Buffer B (20 mM Tris pH 7.9, 150 mM NaCl, 0.25 mM TCEP) and concentrated for crystallization assays to 18 mg/mL by centrifugal ultrafiltration (Millipore). The protein was crystallized using the nanodroplet vapor diffusion method11 with standard JCSG crystallization protocols.6 The crystallization solution contained 2.4 M (NH4)2SO4 and 0.1 M 2-[N-Cyclohexylamino] ethane sulfonic acid (CHES) at pH 9.0. The crystals were indexed in the orthorhombic space group P212121 (Table I).
Data collection: Native diffraction data were collected at Stanford Synchrotron Radiation Laboratory (SSRL, Stanford, CA) on beamline 9-1 using the BLU-ICE12 data-collection environment (Table I). The data set was collected at 100 K using a Quantum 315 charge-coupled device (CCD) detector. Data were integrated and reduced using MOSFLM13 and then scaled with the program SCALA from the CCP4 suite.14 Data statistics are summarized in Table I.
Structure solution and refinement: The structure was determined by molecular replacement using the program MOLREP from the CCP4 suite.14 A homology model based on the FFAS8 alignment between TM1255 and the AspAT from T. thermophilus (PDB code: 1BKG) with a sequence identity of 43% was constructed with the modeling program WHATIF15 and used as a search model. Structure refinement was performed using REFMAC5,14 O,16 and Xfit.17 Refinement statistics are summarized in Table I. The final model includes 2 protein molecules [residues 2–376 for chain A, residues 1–376 plus 8 residues from the N-terminal expression- and purification-tags (-7-KIHHHHHH-0) for chain B], 2 covalently bound PLP molecules (copurified), 10 sulphates, and 641 water molecules in the asymmetric unit. No electron density was observed for residue 377 in both chains and the His-tag in chain A.
Validation and deposition: Analysis of the stereochemical quality of the models was accomplished using the JCSG Validation Central suite, which integrates 7 validation tools: PROCHECK 3.5.4, SFCHECK 4.0, PROVE 2.5.1, ERRAT, WASP, DDQ 2.0, and WHATCHECK. The Validation Central suite is accessible at http://www.jcsg.org. Figure 1B was adapted from an analysis using PDBsum (http://www.biochem.ucl.ac.uk/bsm/pdbsum/) and all others were prepared with PYMOL (DeLano Scientific). Atomic coordinates of the final model and experimental structure factors of TM1255 have been deposited with the PDB and are accessible under the code 1O4S.
Acknowledgements
Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health (National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences).




