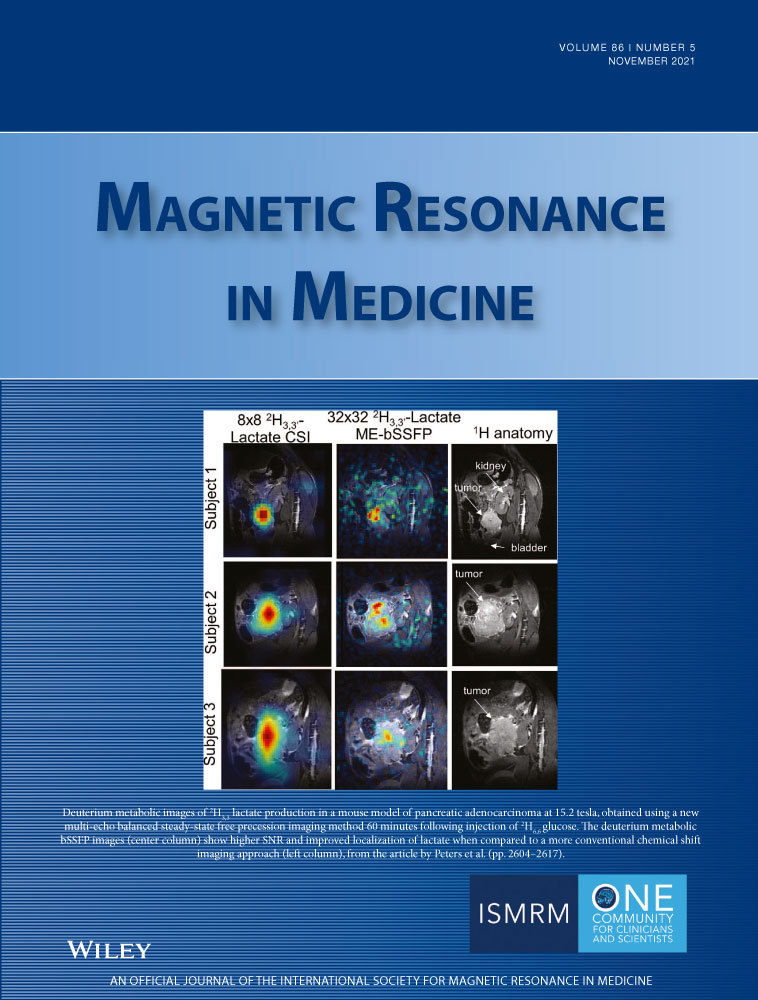
Denoising of hyperpolarized 13C MR images of the human brain using patch-based higher-order singular value decomposition
Yaewon Kim
Department of Radiology and Biomedical Imaging, University of California, San Francisco, California, USA
Search for more papers by this authorHsin-Yu Chen
Department of Radiology and Biomedical Imaging, University of California, San Francisco, California, USA
Search for more papers by this authorAdam W. Autry
Department of Radiology and Biomedical Imaging, University of California, San Francisco, California, USA
Search for more papers by this authorJavier Villanueva-Meyer
Department of Radiology and Biomedical Imaging, University of California, San Francisco, California, USA
Search for more papers by this authorSusan M. Chang
Department of Neurological Surgery, University of California, San Francisco, California, USA
Search for more papers by this authorYan Li
Department of Radiology and Biomedical Imaging, University of California, San Francisco, California, USA
Search for more papers by this authorPeder E. Z. Larson
Department of Radiology and Biomedical Imaging, University of California, San Francisco, California, USA
Search for more papers by this authorJeffrey R. Brender
Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA
Search for more papers by this authorMurali C. Krishna
Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA
Search for more papers by this authorDuan Xu
Department of Radiology and Biomedical Imaging, University of California, San Francisco, California, USA
Search for more papers by this authorDaniel B. Vigneron
Department of Radiology and Biomedical Imaging, University of California, San Francisco, California, USA
Department of Neurological Surgery, University of California, San Francisco, California, USA
Search for more papers by this authorCorresponding Author
Jeremy W. Gordon
Department of Radiology and Biomedical Imaging, University of California, San Francisco, California, USA
Correspondence
Jeremy W. Gordon, Department of Radiology and Biomedical Imaging, University of California San Francisco, 1700 4th St, Byers Hall 102, San Francisco, CA 94158, USA.
Email: [email protected]
Search for more papers by this authorYaewon Kim
Department of Radiology and Biomedical Imaging, University of California, San Francisco, California, USA
Search for more papers by this authorHsin-Yu Chen
Department of Radiology and Biomedical Imaging, University of California, San Francisco, California, USA
Search for more papers by this authorAdam W. Autry
Department of Radiology and Biomedical Imaging, University of California, San Francisco, California, USA
Search for more papers by this authorJavier Villanueva-Meyer
Department of Radiology and Biomedical Imaging, University of California, San Francisco, California, USA
Search for more papers by this authorSusan M. Chang
Department of Neurological Surgery, University of California, San Francisco, California, USA
Search for more papers by this authorYan Li
Department of Radiology and Biomedical Imaging, University of California, San Francisco, California, USA
Search for more papers by this authorPeder E. Z. Larson
Department of Radiology and Biomedical Imaging, University of California, San Francisco, California, USA
Search for more papers by this authorJeffrey R. Brender
Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA
Search for more papers by this authorMurali C. Krishna
Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA
Search for more papers by this authorDuan Xu
Department of Radiology and Biomedical Imaging, University of California, San Francisco, California, USA
Search for more papers by this authorDaniel B. Vigneron
Department of Radiology and Biomedical Imaging, University of California, San Francisco, California, USA
Department of Neurological Surgery, University of California, San Francisco, California, USA
Search for more papers by this authorCorresponding Author
Jeremy W. Gordon
Department of Radiology and Biomedical Imaging, University of California, San Francisco, California, USA
Correspondence
Jeremy W. Gordon, Department of Radiology and Biomedical Imaging, University of California San Francisco, 1700 4th St, Byers Hall 102, San Francisco, CA 94158, USA.
Email: [email protected]
Search for more papers by this authorFunding information
National Institutes of Health, Grant/Award Numbers: P41EB013598, P01CA118816, T32CA151022, P50CA097257; UCSF NICO project
Abstract
Purpose
To improve hyperpolarized 13C (HP-13C) MRI by image denoising with a new approach, patch-based higher-order singular value decomposition (HOSVD).
Methods
The benefit of using a patch-based HOSVD method to denoise dynamic HP-13C MR imaging data was investigated. Image quality and the accuracy of quantitative analyses following denoising were evaluated first using simulated data of [1-13C]pyruvate and its metabolic product, [1-13C]lactate, and compared the results to a global HOSVD method. The patch-based HOSVD method was then applied to healthy volunteer HP [1-13C]pyruvate EPI studies. Voxel-wise kinetic modeling was performed on both non-denoised and denoised data to compare the number of voxels quantifiable based on SNR criteria and fitting error.
Results
Simulation results demonstrated an 8-fold increase in the calculated SNR of [1-13C]pyruvate and [1-13C]lactate with the patch-based HOSVD denoising. The voxel-wise quantification of kPL (pyruvate-to-lactate conversion rate) showed a 9-fold decrease in standard errors for the fitted kPL after denoising. The patch-based denoising performed superior to the global denoising in recovering kPL information. In volunteer data sets, [1-13C]lactate and [13C]bicarbonate signals became distinguishable from noise across captured time points with over a 5-fold apparent SNR gain. This resulted in >3-fold increase in the number of voxels quantifiable for mapping kPB (pyruvate-to-bicarbonate conversion rate) and whole brain coverage for mapping kPL.
Conclusions
Sensitivity enhancement provided by this denoising significantly improved quantification of metabolite dynamics and could benefit future studies by improving image quality, enabling higher spatial resolution, and facilitating the extraction of metabolic information for clinical research.
Open Research
DATA AVAILABILITY STATEMENT
The code and data that support the findings of this study are openly available in hp13C_EPI-hosvd_denoising at https://github.com/ykim-hmtrc/hp13c_EPI-hosvd_denoising.
Supporting Information
| Filename | Description |
|---|---|
| mrm28887-sup-0001-Supinfo.docxWord document, 4.4 MB |
FIGURE S1 Simulated assessment of kPL accuracy and precision. (A) The error images showing the mean percent differences (bias) in kPL estimates between noise-added and noise-free images (A,D,G); the corresponding GL-HOSVD processed and noise-free images (B,E,H); and the TRI processed and noise-free images (C,F,I), calculated from 500 simulated data sets. The SD of the random Gaussian noise contained in the noise-added data (σn = 0.2) is indicated on the left; (B) Mean SEs for the fitted kPL values from 500 noise-added simulated data (A,D,G), and corresponding GL-HOSVD (B,E,H) and TRI images (C,F,I) at different noise levels FIGURE S2 Plot of mean kPL error (%) obtained from denoised kPL maps versus the original SNRauc of lactate. Mean kPL errors calculated from white and gray matter of kPL maps obtained from 500 GL-HOSVD processed data sets simulated with 20%, 30%, and 40%, and additionally 50%, 80%, 100%, and 150% random Gaussian noise, and plotted over the original (non-denoised) SNRauc (SNR of temporally summed metabolic signals) of lactate from the corresponding brain region. The error bars indicate the SD of the mean kPL error FIGURE S3 Simulated assessment of accuracy of under the curve ratio (Lacauc/Pyrauc). Error images showing the mean percent differences (bias) in lactate-to-pyruvate area under the curve ratio estimates between noise-added and noise-free images (A,D,G); the corresponding GL-HOSVD processed and noise-free images (B,E,H); and the TRI processed and noise-free images (C,F,I), calculated from 500 simulated data sets. The SD of the random Gaussian noise contained in the noise-added data is indicated on the left FIGURE S4 Denoising results of GL-HOSVD for the multi-slice, dynamic HP-13C EPI data acquired from a healthy brain volunteer using a multi-channel coil. Multi-slice images from the data presented in Figure 6. The original (Orig.) and denoised (DN) metabolite images from the early and later time points (t = 14 and 38 s, corresponding to the 4th and 12th scans, respectively), are displayed for 5 axial slices. The images have been 0-filled 4-fold for display. The upper window level was adjusted to 55% (pyruvate) or 110% (lactate and bicarbonate) of the maximum intensity voxel in the 5 slices of the denoised images. For anatomic reference, 1H T2-FSE images are shown on the left. Noise reduction for the pyruvate images can be clearly seen in the later time point, and for both time points in the lactate and bicarbonate images FIGURE S5 Representative traces of original and denoised [1-13C]pyruvate, [1-13C]lactate, and [13C]bicarbonate signals from a selected voxel of the healthy volunteer data. The selected voxel for each data set (HV-01, HV-02, and HV-06) is marked on the anatomic 1H image on the left. The original and denoised data points are plotted with blue circles and orange crosses, respectively. The SNRauc for the raw pyruvate, lactate, and bicarbonate data before denoising, respectively, for HV-01, HV-02, and HV-06 are (50, 24, 7), (58, 31, 9), and (20, 12, 3) FIGURE S6 Comparison of kPL and kPB values obtained from the original and denoised in vivo data. (A) Correlation plots of kPL estimates from the raw images and those of the corresponding voxels from the GL-HOSVD denoised images (DN) of the data set HV-01 at 5 different SNRauc ranges of raw lactate signals. (B) Correlation plots of kPB estimates from the raw images and those of the corresponding voxels from the GL-HOSVD denoised images (DN) of the data set HV-01 at 3 different SNRauc ranges of raw bicarbonate signals. The red diagonal lines indicate when the ratio of kPL(raw) to kPL(DN) is 1. The average of the ratios is indicated in each plot FIGURE S7 Comparison of original and denoised metabolite dynamic data. Traces of the original and denoised metabolite signals in arbitrary units (a.u.) from 4 selected voxels, indicated by green boxes in the kPL maps with numbered labels, are shown. For each voxel, pyruvate and lactate data points before (“Orig.”; blue) and after denoising (“DN”; orange) are displayed. The SNRauc for the raw lactate signals are denoted in each plot. In the lactate graphs, the curves from kPL fitting of the original and denoised data are also shown with dashed black lines and solid orange lines, respectively. The corresponding fitted kPL values are plotted in the bottom graphs with error bars representing SDs of the fitted kPL. The fitted curves of the original and denoised lactate signals from voxels no. 1 and 4 showed good agreement whereas the kPL values from the denoised data were determined with higher precision than those from the original data as can be seen from the kPL error bars. From voxels no. 2 and 3 where the lactate signals were relatively low, the initial signal rise was not accurately captured for the original data because of fluctuating initial data points, resulting in high kPL with low precision FIGURE S8 Effect of patch size on the performance of GL-HOSVD. Three different patch sizes (3 × 3, 5 × 5, 7 × 7) were applied in GL-HOSVD to denoise a dynamic series of pyruvate and lactate images with random Gaussian noise σn = 0.4, and kPL maps were calculated from the resulting images. (A-C) Ground truth kPL maps for the whole brain slice, gray matter, and white matter (top to bottom); the kPL maps from the denoised data using the patch size of 3 × 3 (D-F), 5 × 5 (G-I), and 7 × 7 (J-L) for the whole brain slice, gray matter, and white matter (top to bottom). As the patch size increases, the kPL maps contain less noise but lose finer details, as manifested by the blurred edges TABLE S1 Summary of metabolite SNRauc; spatial coverage in kPL and kPB maps; and the mean voxel-wise kPL and kPB values from the original (Orig.) and denoised (DN) dynamic HP-13C EPI images of the brain acquired from 6 healthy volunteers |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
REFERENCES
- 1Ardenkjær-Larsen JH, Fridlund B, Gram A, et al. Increase in signal-to-noise ratio of >10,000 times in liquid-state NMR. PNAS. 2003; 100: 10158-10163.
- 2Wang ZJ, Ohliger MA, Larson PEZ, et al. Hyperpolarized 13C MRI: state of the art and future directions. Radiology. 2019; 291: 273-284.
- 3Kurhanewicz J, Vigneron DB, Ardenkjaer-Larsen JH, et al. Hyperpolarized 13C MRI: path to clinical translation in oncology. Neoplasia. 2019; 21: 1-16.
- 4Park I, Larson PEZ, Zierhut ML, et al. Hyperpolarized 13C magnetic resonance metabolic imaging: application to brain tumors. Neuro-Oncology. 2010; 12: 133-144.
- 5Park JM, Josan S, Grafendorfer T, et al. Measuring mitochondrial metabolism in rat brain in vivo using MR spectroscopy of hyperpolarized [2-13C]pyruvate: measuring mitochondrial metabolism In brain using [2- 13C]pyruvate. NMR Biomed. 2013; 26: 1197-1203.
- 6Mishkovsky M, Anderson B, Karlsson M, et al. Measuring glucose cerebral metabolism in the healthy mouse using hyperpolarized 13C magnetic resonance. Sci Rep. 2017; 7: 11719.
- 7Mishkovsky M, Comment A, Gruetter R. In Vivo detection of brain krebs cycle intermediate by hyperpolarized magnetic resonance. J Cereb Blood Flow Metab. 2012; 32: 2108-2113.
- 8Najac C, Ronen SM. MR molecular imaging of brain cancer metabolism using hyperpolarized 13C magnetic resonance spectroscopy. Top Magn Reson Imaging. 2016; 25: 187-196.
- 9Park I, Larson PEZ, Gordon JW, et al. Development of methods and feasibility of using hyperpolarized carbon-13 imaging data for evaluating brain metabolism in patient studies: hyperpolarized carbon-13 metabolic imaging of patients with brain tumors. Magn Reson Med. 2018; 80: 864-873.
- 10Chung BT, Chen H-Y, Gordon J, et al. First hyperpolarized [2-13C]pyruvate MR studies of human brain metabolism. J Magn Reson. 2019; 309: 2-13.
- 11Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008; 134: 703-707.
- 12Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012; 21: 297-308.
- 13Crane JC, Gordon JW, Chen H-Y, et al. Hyperpolarized 13C MRI data acquisition and analysis in prostate and brain at University of California, San Francisco. NMR Biomed. 2021; 34(5):e4280.
- 14Park I, Larson PEZ, Gordon JW, et al. Development of methods and feasibility of using hyperpolarized carbon-13 imaging data for evaluating brain metabolism in patient studies. Magn Reson Med. 2018; 80: 864-873.
- 15Miloushev VZ, Granlund KL, Boltyanskiy R, et al. Metabolic imaging of the human brain with hyperpolarized 13C pyruvate demonstrates 13C lactate production in brain tumor patients. Cancer Res. 2018; 78: 3755-3760.
- 16Autry AW, Gordon JW, Chen H-Y, et al. Characterization of serial hyperpolarized 13C metabolic imaging in patients with glioma. Neuroimage Clin. 2020; 27: 102323.
- 17Lee CY, Soliman H, Geraghty BJ, et al. Lactate topography of the human brain using hyperpolarized 13C-MRI. Neuroimage. 2020; 204: 116202.
- 18Grist JT, McLean MA, Riemer F, et al. Quantifying normal human brain metabolism using hyperpolarized [1–13C]pyruvate and magnetic resonance imaging. Neuroimage. 2019; 189: 171-179.
- 19Gordon JW, Hansen RB, Shin PJ, Feng Y, Vigneron DB, Larson PEZ. 3D hyperpolarized C-13 EPI with calibrationless parallel imaging. J Magn Reson. 2018; 289: 92-99.
- 20Tang S, Bok R, Qin H, et al. A metabolite-specific 3D stack-of-spiral bSSFP sequence for improved lactate imaging in hyperpolarized [1-13C]pyruvate studies on a 3T clinical scanner. Magn Reson Med. 2020; 84: 1113-1125.
- 21Deh K, Granlund KL, Eskandari R, Kim N, Mamakhanyan A, Keshari KR. Dynamic volumetric hyperpolarized 13C imaging with multi-echo EPI. Magn Reson Med. 2021; 85(2): 978-986.
- 22Chen H, Larson PEZ, Gordon JW, et al. Technique development of 3D dynamic CS-EPSI for hyperpolarized 13C pyruvate MR molecular imaging of human prostate cancer. Magn Reson Med. 2018; 80: 2062-2072.
- 23Larson PEZ, Kerr AB, Chen AP, et al. Multiband excitation pulses for hyperpolarized 13C dynamic chemical-shift imaging. J Magn Reson. 2008; 194: 121-127.
- 24Gordon JW, Autry AW, Tang S, et al. A variable resolution approach for improved acquisition of hyperpolarized 13C metabolic MRI. Magn Reson Med. 2020; 84: 2943-2952.
- 25Autry AW, Gordon JW, Carvajal L, et al. Comparison between 8- and 32-channel phased-array receive coils for in vivo hyperpolarized 13C imaging of the human brain. Magn Reson Med. 2019; 82: 833-841.
- 26Sánchez-Heredia JD, Olin RB, McLean MA, et al. Multi-site benchmarking of clinical 13C RF coils at 3T. J Magn Reson. 2020; 318: 106798.
- 27Manjon J, Carbonellcaballero J, Lull J, Garciamarti G, Martibonmati L, Robles M. MRI denoising using non-local means. Med Image Anal. 2008; 12: 514-523.
- 28Meurée C, Maurel P, Ferré J-C, Barillot C. Patch-based super-resolution of arterial spin labeling magnetic resonance images. Neuroimage. 2019; 189: 85-94.
- 29Maggioni M, Katkovnik V, Egiazarian K, Foi A. Nonlocal transform-domain filter for volumetric data denoising and reconstruction. IEEE Trans on Image Process. 2013; 22: 119-133.
- 30Wood JC, Johnson KM. Wavelet packet denoising of magnetic resonance images: importance of Rician noise at low SNR. Magn Reson Med. 1999; 41: 631-635.
10.1002/(SICI)1522-2594(199903)41:3<631::AID-MRM29>3.0.CO;2-Q CAS PubMed Web of Science® Google Scholar
- 31Ma C, Clifford B, Liu Y, et al. High-resolution dynamic 31P-MRSI using a low-rank tensor model: 31P-spice. Magn Reson Med. 2017; 78: 419-428.
- 32Liu Y, Ma C, Clifford B, Lam F, Johnson C, Liang Z-P. Improved low-rank filtering of magnetic resonance spectroscopic imaging data corrupted by noise and B0 field inhomogeneity. IEEE Trans Biomed Eng. 2015; 63(4): 841.
- 33Nguyen HM, Peng X, Do MN, Liang Z-P. Denoising MR spectroscopic imaging data with low-rank approximations. IEEE Trans Biomed Eng. 2013; 60: 78-89.
- 34Zhang X, Peng J, Xu M, et al. Denoise diffusion-weighted images using higher-order singular value decomposition. Neuroimage. 2017; 156: 128-145.
- 35Zhang X, Xu Z, Jia N, et al. Denoising of 3D magnetic resonance images by using higher-order singular value decomposition. Med Image Anal. 2015; 19: 75-86.
- 36De Lathauwer L, De Moor B, Vandewalle J. A multilinear singular value decomposition. SIAM J Matrix Anal Appl. 2000; 21: 1253-1278.
- 37Rajwade A, Rangarajan A, Banerjee A. Image denoising using the higher order singular value decomposition. IEEE Trans Pattern Anal Mach Intell. 2013; 35: 849-862.
- 38Brender JR, Kishimoto S, Merkle H, et al. Dynamic imaging of glucose and lactate metabolism by 13C-MRS without hyperpolarization. Sci Rep. 2019; 9: 3410.
- 39Chen H, Autry AW, Brender JR, et al. Tensor image enhancement and optimal multichannel receiver combination analyses for human hyperpolarized 13C MRSI. Magn Reson Med. 2020; 84: 3351-3365.
- 40Donoho DL, Johnstone IM. Ideal spatial adaptation by wavelet shrinkage. Biometrika. 1994; 81: 425-455.
- 41Collins DL, Zijdenbos AP, Kollokian V, et al. Design and construction of a realistic digital brain phantom. IEEE Trans Med Imaging. 1998; 17: 463-468.
- 42Larson PEZ, Chen H-Y, Gordon JW, et al. Investigation of analysis methods for hyperpolarized 13C-pyruvate metabolic MRI in prostate cancer patients: hyperpolarized pyruvate prostate cancer analysis methods. NMR Biomed. 2018; 31:e3997.
- 43Mammoli D, Gordon J, Autry A, et al. Kinetic modeling of hyperpolarized carbon-13 pyruvate metabolism in the human brain. IEEE Trans Med Imaging. 2020; 39: 320-327.
- 44Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process. 2004; 13: 600-612.
- 45Gordon JW, Chen H-Y, Autry A, et al. Translation of carbon-13 EPI for hyperpolarized MR molecular imaging of prostate and brain cancer patients. Magn Reson Med. 2019; 81: 2702-2709.
- 46Zhu Z, Zhu X, Ohliger MA, et al. Coil combination methods for multi-channel hyperpolarized 13C imaging data from human studies. J Magn Reson. 2019; 301: 73-79.
- 47Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. 2001; 5: 143-156.
- 48Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001; 20: 45-57.