Gene correction of the CLN3 c.175G>A variant in patient-derived induced pluripotent stem cells prevents pathological changes in retinal organoids
Abstract
Background
Mutations in CLN3 cause Batten disease, however non-syndromic CLN3 disease, characterized by retinal-specific degeneration, has been also described. Here, we characterized an induced pluripotent stem cell (iPSC)-derived disease model derived from a patient with non-syndromic CLN3-associated retinopathy.
Methods
Patient-iPSC, carrying the 1 kb-deletion and c.175G>A variants in CLN3, coisogenic iPSC, in which the c.175G>A variant was corrected, and control iPSC were differentiated into neural retinal organoids (NRO) and cardiomyocytes. CLN3 transcripts were analyzed by Sanger sequencing. Gene expression was characterized by qPCR and western blotting. NRO were characterized by immunostaining and electron microscopy.
Results
Novel CLN3 transcripts were detected in adult human retina and control-NRO. The major transcript detected in patient-NRO displayed skipping of exons 2 and 4–9. Accumulation of subunit-C of mitochondrial ATPase (SCMAS) protein was demonstrated in patient-derived cells. Photoreceptor progenitor cells in patient-NRO displayed accumulation of peroxisomes and vacuolization of inner segments. Correction of the c.175G>A variant restored CLN3 mRNA and protein expression and prevented SCMAS and inner segment vacuolization.
Conclusion
Our results demonstrate the expression of novel CLN3 transcripts in human retinal tissues. The c.175G>A variant alters splicing of the CLN3 pre-mRNA, leading to features consistent with CLN3 deficiency, which were prevented by gene correction.
1 INTRODUCTION
Juvenile neuronal ceroid lipofuscinosis (JNCL), also known as Batten Disease, is a heterogeneous neurodegenerative condition associated with declining cognitive and motor function, progressive visual impairment, cerebellar atrophy and premature death (Mitchison et al., 1998; Rapola, 1993; Santavuori, 1988; Santavuori et al., 2000). A 1 kb deletion in the ceroid lipofuscinosis neuronal 3 (CLN3) gene was identified in 1995 as a cause of JNCL (Lerner et al., 1995). More recently, isolated retinal degeneration without neurological or mental impairment has been associated with novel missense mutations in CLN3 (Wang et al., 2014) and detailed descriptions of the retinal phenotype and natural history of non-syndromic CLN3-associated retinopathy have been described (Chen et al., 2019; Ku et al., 2017). Previous studies have shown that approximately 74% of patients with CLN3 disease carry a homozygous 966 bp genomic deletion (c.461–280_677+382del966, also known as the 1 or 1.02 kb deletion) that removes exons 7 and 8 (Jarvela et al., 1997). The remaining cases were heterozygous for the 1 kb deletion and a missense, nonsense, indel, intronic or splice-site mutation (de los Reyes et al., 2004; Ku et al., 2017; Mole et al., 2005).
The CLN3 gene encodes a ubiquitously expressed (Margraf et al., 1999; Oetjen et al., 2016), glycosylated integral transmembrane protein of 438 amino acids with six transmembrane domains and cytosolic N- and C-termini (Nugent et al., 2008; Ratajczak et al., 2014). CLN3 protein has been localized to the Golgi network in fibroblasts (Persaud-Sawin et al., 2004), lysosomes in HeLa cells (Jarvela et al., 1998), synaptic vesicles in transfected rat PC6-3 cells (Haskell et al., 2000), synaptosomes in mouse brain (Luiro et al., 2001), and mitochondria in mouse Müller cells (Katz et al., 1997), however, the precise cellular functions of CLN3 remain unclear. The expanding profile of CLN3 protein interactions includes associations with trafficking adaptors (AP-1 and AP-3), microtubule binding proteins (Hook1) and the large family of Rab GTPases suggesting a role in endocytosis and endocytic membrane trafficking (Getty & Pearce, 2011; Luiro et al., 2004). CLN3 also has a role in protein trafficking through an intracellular pathway connecting the Golgi apparatus with endosomes, autophagosomes, lysosomes, and the plasma membrane (Cotman & Staropoli, 2012; Persaud-Sawin et al., 2004). CLN3 deficiency has been associated with reduced cellular growth rates and susceptibility to apoptosis (Mao et al., 2015; Persaud-Sawin et al., 2002). In homozygous Cln3∆ex7/8 mice, microglia presented enhanced caspase-1 activities (Xiong & Kielian, 2013) while astrocytes showed a significant decline in glutamine synthetase (Burkovetskaya et al., 2014), which all lead to the disruption of glutamate homeostasis. Accumulation of subunit c of mitochondrial ATP synthase (SCMAS) was observed in the cerebellum of homozygous Cln3∆7/8 mice (Cao et al., 2011), as well as in neural cells differentiated from JNCL patient-derived induced pluripotent stem cells (iPSC) (Lojewski et al., 2014; Ryazantsev et al., 2007).
We previously described the clinical characteristics of a patient with isolated retinal degeneration due to compound heterozygous mutations (c.175G>A and the 1 kb deletion) in CLN3 (NM_000086.2) (Chen et al., 2019). We further described the generation of iPSC from her dermal fibroblasts, and gene editing to produce coisogenic control iPSC in which the c.175G>A mutation was corrected (Zhang et al., 2018). Here, we have expanded on this work by differentiating these iPSC into neural retinal organoids (NRO) and cardiomyocytes (CM) to determine whether (a) the c.175G>A point mutation affects CLN3 pre-mRNA splicing and protein production, and (b) the patient's NRO and CM display molecular and histopathological features of CLN3 deficiency. Our results demonstrate the c.175G>A mutation leads to altered CLN3 splicing, and that patient iPSC, NRO and CM accumulate SCMAS, while gene-corrected controls do not. Additionally, we report the identification of novel CLN3 transcripts expressed in adult human retina and NRO.
2 MATERIALS AND METHODS
Collection of patient samples and generation of iPSC was approved by the Human Research Ethics Committee of The University of Western Australia (RA/4/1/7916). Written consent was obtained from the patient and all procedures were carried out in accordance with the requirements of the National Health & Medical Research Council of Australia and the Declaration of Helsinki.
2.1 Cell culture and differentiation
In this study, we utilized iPSC derived from a female patient with non-syndromic CLN3 disease carrying the c.175G>A and 1 kb deletion variants in CLN3. The patient's clinical phenotype and genetic diagnosis was recently described (Chen et al., 2019). The generation and characterization of the patient iPSC line LEIi004-A and the coisogenic control line LEIi004-A-1, in which the c.175G>A variant was corrected by CRISPR/Cas9 gene editing, has been previously described (Zhang et al., 2018). Briefly, dermal fibroblasts derived from a patient skin biopsy were reprogrammed using the Episomal iPSC Reprogramming Plasmid kit (SC900A-1, System Biosciences) according to the manufacturer's instructions. For controls, we utilized a commercially available human iPSC line (ThermoFisher, Cat#A18945). Human iPSC were cultured in feeder-free conditions, on geltrex (ThermoFisher) coated culture plates in TeSR-E1 medium (Stem Cell Technologies). For directed differentiation into iPSC-neural retinal organoids (NRO) we followed a previously published protocol (Mellough et al., 2015). Briefly, iPSCs were dissociated into small pieces by 5 nM EDTA (ThermoFisher) buffer prior to suspension cultivation in the DMEM/F-12 medium (ThermoFisher) with decreasing concentrations of KOSR (20% for the first 5 days, 15% until day 12, 10% until day 35) containing l-glutamine, NEAA, B27 and IGF-1 (ThermoFisher). For cardiomyocyte (CM) differentiation, we used the PSC Cardiomyocyte Differentiation Kit (ThermoFisher, Cat# A2921201), according to the manufacturer's instructions.
2.2 RNA isolation and reverse transcription
Total mRNA was isolated using TRIzol reagent. NRO from one well of a 6-well plate were suspended in 500 μl TRIzol reagent (ThermoFisher). After homogenization and incubation at room temperature for 5 min, samples were centrifuged at 12,000× g for 10 min at 4°C. The supernatants were then collected and 0.1 ml of chloroform was added to each sample and mixed for 30 s using a vortex mixer. Samples were then incubated at room temperature for 5 min and centrifuged at 12,000× g for 15 min at 4°C. The supernatant was removed and the pellet was washed twice with 1 ml of 75% ethanol and centrifuged at 12,000× g for 10 min at 4°C. The RNA pellet was air dried at room temperature and resuspended in 20 μl DEPC water (ThermoFisher). The cDNA was synthesized using the RT2 First Strand Kit (Qiagen), according to the manufacturer's instructions.
2.3 PCR and cloning
CLN3 cDNA fragments were amplified with Q5 High-Fidelity Polymerase (New England Biolabs) using primer pairs targeting specific CLN3 exons (Table S1). After purification with PCR Clean-up System (Promega), fragments were cloned into pJET-Blunt Cloning Vector (ThermoFisher). Twelve clones for each were picked up and screened by PCR using primers for the vector (Forward: CGACTCACTATAGGGAGAGCGGC; Reverse: AAGAACATCGATTTTCCATGGCAG). The positive clones containing CLN3 cDNA fragments were selected for culture in LB medium overnight at 37°C. Plasmids were extracted by QIAprep spin Miniprep Kit (Qiagen) and then sent for Sanger sequencing (Australian Genome Research Facility).
2.4 Quantitative PCR
Quantitative polymerase chain reactions (qPCR) were performed using the RT2 SYBR Green qPCR Mastermix (Qiagen) and the CFX96TM Real-Time System (BioRad). Primers used for qPCR are listed in Table S2. Data were analyzed using the ΔΔCT method. Gene expression values were normalized to GAPDH expression and expressed as fold changes compared with undifferentiated iPSC.
2.5 Western blotting
Samples were collected and protein was extracted on ice using RIPA (ThermoFisher) with 1% Protease Inhibitor Cocktail (Sigma) for half an hour, centrifuged at 15,000× g at 4°C for 20 min and the supernatant was collected. Protein concentration was measured by the Bradford (Biorad) method and the sample was boiled with 4X loading dye (ThermoFisher) and 19X 2-Mercaptoethanol (ThermoFisher). Samples were loaded and run on ice (ThermoFisher, Cat#NP0321BOX) at 80 V for the stacking gel and 100 V for the resolving gel, then transferred to a PVDF membrane for 1.5 hr at 300 mA. The membrane was blocked with 5% BSA/TBS for one hour. Primary antibodies diluted with 5% BSA/TBST were applied at 4°C overnight. Secondary antibodies were applied for 2 hr at room temperature. Antibodies used for western blotting include rabbit anti-CLN3 (1:1000, Invitrogen Cat# PA549382), rabbit anti-ATP-Synthase C (SCMAS) (1:1000, Abcam/ab181243), and mouse anti-GAPDH (1:2000, Abcam Cat# ab125247). Two secondary antibodies from LI-COR Odyssey are used in this study: IRDye 680RD (goat anti-mouse) and IRDye 800RD (goat anti-rabbit). Images were taken using an infrared imaging system (LI-COR Odyssey). For quantification of protein expression, the background was subtracted from each band and normalized to the respective control GAPDH band using ImageJ software (https://imagej.nih.gov/ij/).
2.6 Transmission electron microscopy (TEM) analysis
Retinal organoids were fixed after 35 days of differentiation with 4% paraformaldehyde, 2% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4 for 24 hr at 4°C and then post-fixed in 1% osmium tetroxide in the same buffer for 6 min (2 min on, 2 min off, and 2 min on) using the BioWave Processing System (PELCO). Samples were dehydrated in a graded series of ethanol and acetone, followed by resin infiltration. Resin included Procure 812, Araldite 502, DDSA and BDMA. Ultrathin (100 nm) sections were scanned with a JEM2100 electron microscope (JEOL).
2.7 Statistical analysis
Statistical analysis was performed using GraphPad Prism (GraphPad Software). All values were expressed as mean ± SEM. Statistical significance was calculated by t-test. For the analyses in this study, a p value <.05 was considered statistically significant.
3 RESULTS
Patient-derived dermal fibroblasts were previously reprogrammed to produce the LEIi004-A patient iPSC line, carrying biallelic mutations in CLN3 (c.175G>A and the 1 kb deletion). The c.175G>A mutation was corrected in patient iPSC by CRISPR/Cas9 gene editing to produce the LEIi004-A-1 coisogenic control iPSC line. Both LEIi004-A and LEIi004-A-1 iPSC expressed pluripotency markers, displayed normal karyotypes, and demonstrated trilineage differentiation potential (Zhang et al., 2018). To prevent recognition and re-cutting of Cas9 targeting sites during gene editing, two synonymous mutations were introduced into the repair template (Figure S1). In silico splice site analysis (Human Splicing Finder 3.1, http://www.umd.be/HSF/) predicted minimal impact of these additional mutations on CLN3 splicing (Figure S2). For healthy control iPSC, we utilized a commercially available iPSC line (HuiPSC).
LEIi004-A, LEIi004-A-1, and HuiPSC control iPSC were differentiated into NRO for 10–16 weeks and CM for 4 weeks. After 10 weeks, differentiating NRO from all three lines displayed upregulation of six retinal markers, including PAX6, RPE65, CHX10, OTX2, RCVRN, and NRL (Figure S3A). After 4 weeks, differentiating CM from all three lines displayed spontaneous contraction (Movies V1–V3) and upregulation of four cardiac markers (NKX2.5, GATA4, MYH6 and TNNT2) compared with undifferentiated iPSC (Figure S3B). The pluripotency marker OCT4 was downregulated in cardiomyocytes, compared with iPSC. These results demonstrate successful generation of retinal and cardiac tissues from iPSC.
3.1 CLN3 transcript analysis in adult retina and retinal organoids
CLN3 transcripts were amplified from adult human retina and 16-week NRO cultures using primers targeting exon 1 and exon 15 (Figure 1a). Sanger sequencing demonstrated expression of several CLN3 transcript variants in adult human retinal tissue, the largest (≈1.3 kb) of which corresponded to transcript variant 1/2, while the smallest (≈0.55 kb) corresponded to a novel variant displaying exclusion of exons 3–9 and 11–12 (Figure 1b). Additional bands (approximately 0.8 and 1 kb) were observed, however, we were unable to purify these for cloning and sequencing due to their low abundance.
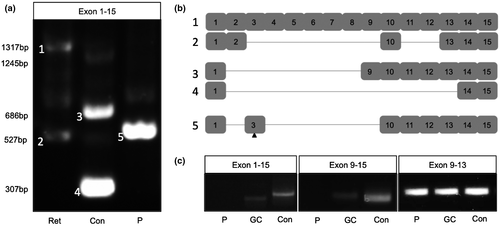
NRO derived from the HuiPSC control line expressed a different subset of transcript variants to those amplified from adult human retina. The largest of these was smaller (≈1.2 kb) than the full-length transcript present in adult human retina, indicating that an alternative transcript may be utilized in the developing retina (Figure 1a). The two most abundant products detected in the control NRO were derived from novel transcripts that displayed exclusion of exons 2–8 (≈0.7 kb) or exons 2–13 (≈0.3 kb) (Figure 1b). Two additional, low intensity bands were detected (≈0.55 and ≈0.9 kb) in control NRO. Together, these results demonstrate the presence of novel alternatively spliced CLN3 transcripts in adult retinal tissues and retinal organoid cultures.
In contrast with control NRO, full length CLN3 mRNA was undetectable in 16-week NRO derived from LEIi004-A patient iPSC. Instead, two PCR products (≈0.6 and ≈0.8 kb) were amplified (Figure 1a). The smaller product displayed exclusion of exons 2 and 4–9 and carried the c.175G>A mutation in exon 3 (Figure 1b), while the larger transcript was present at very low levels. These results demonstrate altered splicing of CLN3 mRNA produced from the c.175G>A allele.
3.2 Gene correction of the c.175G>A variant restores CLN3 transcripts
To further examine the effect of the c.175G>A variant on CLN3 splicing, we performed RT-PCR screening of 10-week NRO derived from the LEIi004-A patient iPSC line, the LEIi004-A-1 gene corrected patient iPSC line and the control iPSC line. Using primers targeting exons 1 and 15, we detected a 0.55 kb band in HuiPSC control NRO, which may correspond to similar sized bands detected in 16-week NRO and adult human retina. No CLN3 transcript was amplified from LEIi004-A patient NRO using these primers, however, correction of the c.175G>A mutation in LEIi004-A-1 resulted in restoration of a slightly smaller CLN3 transcript in NRO. Similarly, using primers targeting exons 9 and 15, a 0.49 kb band, corresponding to full-length CLN3, was detected in HuiPSC control and gene-corrected LEIi004-A-1 NRO, but not in uncorrected LEIi004-A patient NRO (Figure 1c). Together, these results demonstrated that correction of the c.175G>A mutation restored expression of CLN3 transcripts in patient NRO. Interestingly, using primers targeting exons 9 and 13, we detected bands in all lines tested (Figure 1c), suggesting that alternatively spliced CLN3 transcripts lacking exon 15 are produced in early patient NRO.
3.3 CLN3 protein expression in patient iPSC, cardiomyocytes, and retinal organoids
CLN3 protein expression was examined in iPSC, NRO (5-week), and CM (4-week) protein samples by western blotting using an antibody directed at the central portion of the CLN3 protein, encoded by exons 9–11. A CLN3 immunopositive band approximately 50 kDa in size was detected in HuiPSC, LEIi004-A, and LEIi004-A-1 iPSC (Figure 2). LEIi004-A patient iPSC expressed lower levels of CLN3 protein, however, correction of the c.175G>A mutation restored CLN3 protein expression to control levels.
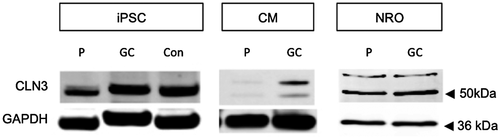
Two CLN3 immunopositive bands approximately 48 and 52 kDa in size were detected in NRO and CM derived from patient LEIi004-A and gene-corrected LEIi004-A-1 iPSC (Figure 2). CLN3 protein expression was reduced in LEIi004-A patient CM compared with gene-corrected LEIi004-A-1 CM (Figure 2). In contrast, patient and gene-corrected NRO expressed similar levels of CLN3 protein.
3.4 Inner segment pathology in patient photoreceptor progenitor cells
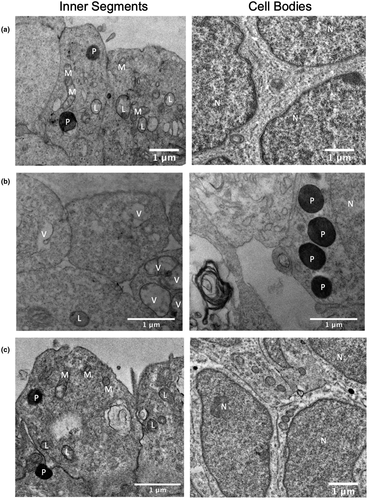
Histological analysis of NRO by TEM demonstrated formation of inner segments by photoreceptor progenitor cells (PPCs) after 5 weeks of differentiation. On the surface of control NRO, PPCs formed tightly packed inner segments containing mitochondria, lysosomes, and occasional peroxisomes. In the developing outer nuclear layer, PPC nuclei were tightly clustered and separated by a thin band of cytoplasm (Figure 3a). In contrast, uncorrected patient NRO formed inner segments largely devoid of identifiable organelles and instead displayed vacuolization. Additionally, PPCs were less tightly packed, with extracellular voids evident around cell bodies and peroxisome accumulation in the cytoplasm (Figure 3b). Gene-corrected patient NRO was similar to controls, with normal inner segment and cell body ultrastructure (Figure 3c).
3.5 Subunit c of mitochondrial ATP synthase accumulation
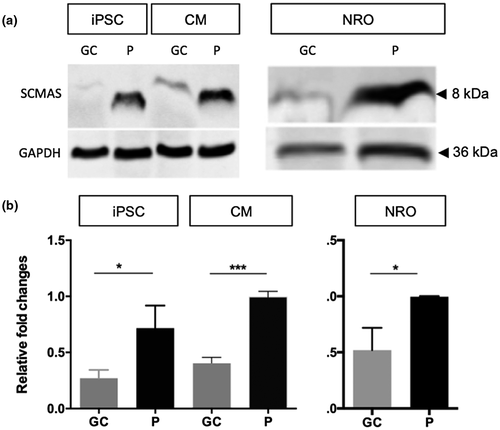
Since CLN3 deficiency has previously been associated with defective mitochondrial autophagy (Cotman et al., 2002; Puranam et al., 1999), we measured the levels of subunit c of mitochondrial protein ATP synthase (SCMAS) protein in uncorrected and gene-corrected patient iPSC-derived cells by western blotting. LEIi004-A patient iPSC and CM displayed significantly increased SCMAS protein levels, compared with gene-corrected LEIi004-A-1 iPSC (p = .0235) and CM (p = .0002). The apparent molecular weight of SCMAS was reduced in uncorrected patient iPSC and CM (Figure 4a). Similar results were obtained in the NRO cultures, with significantly increased SCMAS protein observed in LEIi004-A patient NRO compared with gene-corrected LEIi004-A-1 NRO (p = .0277, Figure 4b). Together, these results demonstrate the accumulation of SCMAS in CLN3Δkb/c.175G>A patient-derived cells and that correction of the c.175G>A variant prevented this accumulation.
4 DISCUSSION
In this study, we investigated the molecular consequences of compound heterozygous CLN3 mutations in a patient with non-syndromic CLN3 disease using patient-derived iPSC. We demonstrated the presence of novel CLN3 transcripts in adult human retina and control NRO. We further demonstrated altered splicing of CLN3 transcripts in patient NRO, which was associated with accumulation of SCMAS protein, mislocalization of peroxisomes and vacuolization of inner segments. Patient iPSC and CM carrying the c.175G>A and 1 kb deletion variants displayed reduced levels of CLN3 protein compared with control cultures. Correction of the c.175G>A variant restored CLN3 mRNA and protein expression, prevented accumulation of SCMAS, and reduced vacuolization of photoreceptor inner segments.
4.1 Alternative splicing of CLN3 in human retinal cells
Previous studies in JNCL patient dermal fibroblasts homozygous for the 1 kb deletion demonstrated the presence of two alternatively spliced CLN3 transcripts. One of these transcripts displayed splicing of exon 6 to exon 9, producing a frameshift mutation and downstream premature termination codon (PTC) in the CLN3 coding sequence. The other transcript displayed splicing of exon 6 to exon 10, resulting in an in-frame deletion of CLN3 coding sequence, predicted to produce a 328 amino acid, internally truncated protein approximately 36 kDa in size. The predicted protein is lacking the second luminal loop, the fourth membrane spanning domain and the third cytosolic loop, the latter of which contains one of the lysosomal targeting regions. Knockdown of these alternative CLN3 transcripts in JNCL patient dermal fibroblasts increased lysosomal size, suggesting translation of a truncated CLN3 protein that retains some lysosomal functions (Kitzmuller et al., 2008). However, it remains unclear whether the truncated CLN3 protein is expressed in human retinal and neural cells and to what extent this mutant protein contributes to CLN3 disease in different tissues.
In a previous study reported by Lojewski et al. (Lojewski et al., 2014), iPSC and iPSC-derived neural progenitor cells were derived from four patients with JNCL, three of which carried homozygous 1 kb deletions, with the fourth carrying compound heterozygous point mutations. Amplification of CLN3 transcripts using primers targeting the 5′ and 3′ UTRs of CLN3 (exons 1 and 15) demonstrated the expression of multiple CLN3 transcript variants in iPSC and neural progenitor cells derived from both control subjects and JNCL patients (Lojewski et al., 2014). Although the sequences of these transcripts were not determined, the results suggest that complex splicing of the CLN3 pre-mRNA could lead to the expression of a range of novel CLN3 transcript variants. In the present study, we detected several CLN3 transcript variants in adult human retinal tissue by RT-PCR, and further identified a novel transcript in which exons 3–9 and 11–12 were skipped (Figure 1a,b). Multiple CLN3 transcripts were also detected in control NRO, with a different profile of products than that observed in adult human retina. The two major CLN3 products detected in 16-week control NRO displayed skipping of exons 2–8 or exons 2–13 (Figure 1a,b). Together, our results indicate alternative splicing of CLN3 pre-mRNA in human retinal cells results in the expression of novel CLN3 transcript variants that differ at distinct developmental timepoints.
Interestingly, the novel transcripts identified in adult retinal tissues and control NRO were all predicted to introduce a frameshift in the coding sequence, resulting in downstream PTCs. Transcripts containing PTCs are often degraded by nonsense-mediated decay (NMD), however, they may escape this process depending on the precise context of the PTC (Cotman et al., 2002). On the other hand, at least two alternative transcriptional start sites have been identified in CLN3 transcript variants 4 (NM_001286105.2), 5 (NM_001286109.2), and 6 (NM_001286110.2), resulting in the translation of alternative CLN3 protein isoforms (battenin isoforms c, d, and e respectively). There are currently more than 60 CLN3 transcript variants listed in the Ensembl database (PMID: 29155950), many of which are predicted to be degraded by NMD. Given the apparent complexity of CLN3 splicing, it is possible that some of these transcripts contain novel alternative start codons that preserve the reading frame of the coding sequence.
4.2 The c.175G>A variant leads to altered splicing of CLN3 pre-mRNA
In silico analysis using Human Splicing Finder 3.1 predicted the c.175G>A mutation would alter splicing of the CLN3 pre-mRNA due to disruption of a group of exonic splicing enhancer sites (ESEs) (Figure S2). In our previous study, CLN3c.175G>A/Δ1kb patient leukocytes displayed significantly reduced CLN3 mRNA levels and expressed two novel transcripts lacking exon 3 and exons 3–5 (Chen et al., 2019). In the present study, we demonstrated altered splicing of transcripts derived from the CLN3c.175G>A allele in patient-derived retinal cells. The 16-week patient NRO did not express the two novel CLN3 transcript variants detected in control NRO (Figure 1a). Similarly, the low abundance of 1.2 and 0.55 kb products detected in control NRO were absent from patient NRO samples. Instead, the major transcript expressed in patient NRO was found to originate from the CLN3c.175G>A allele and displayed skipping of exons 2 and 4–9 (Figure 1a,b). Translation initiation from any of the three known CLN3 translational start sites was predicted to result in a coding sequence frameshift and the introduction of a PTC.
To confirm the effect of the c.175G>A variant on pre-mRNA splicing, we performed RT-PCR screening for CLN3 transcripts in 10-week NRO derived from control, patient and gene-corrected iPSC. CLN3 transcripts containing exons 1 and 15 or 9 and 15 were detected in control NRO, but not in LEIi004-A patient NRO. Correction of the c.175G>A mutation restored expression of these CLN3 transcripts in coisogenic LEIi004-A-1 NRO. Interestingly, using primers targeting exons 9–13, we detected correctly sized amplicons in 10-week NRO of all genotypes, suggesting that CLN3 transcripts with alternative 3’ exon selection may be present.
4.3 Expression of CLN3 proteins
Collectively, the 6 CLN3 transcript variants listed in GenBank encode 5 CLN3 protein isoforms (a–e). Transcript variants 1 and 2 encode the same 47.63 kDa isoform (isoform a), while transcript variant 3 encodes a 45.16 kDa protein (isoform b). The remaining variants encode smaller proteins of 37.03 (c), 39.17 (d), and 41.64 (e) kDa. Using an antibody directed at CLN3 protein sequences encoded by exons 9–11, which are present in all 5 CLN3 protein isoforms, we detected two CLN3 immunopositive bands approximately 45 and 50 kDa in size in both patient and control cells. CLN3 protein levels were reduced in patient-derived LEIi004-A iPSC and CM cultures compared with gene-corrected LEIi004-A-1 or HuiPSC control cultures, indicating correction of the c.175G>A mutation was sufficient to restore CLN3 protein expression in these cells. Unexpectedly, CLN3 protein expression was similar in 10-week patient NRO and gene-corrected patient NRO. Since CLN3 proteins are subject to a number of post-translational modifications (Jarvela et al., 1998), it remains unclear which isoforms contribute to the 48 and 52 kDa bands detected in NRO and CM cultures. While we did not detect CLN3 transcripts containing exons 1–15 or 9–15 in 10-week patient NRO, transcripts containing exons 9–13 were present. Therefore, we propose that the CLN3 proteins detected in 10-week patient and gene corrected NRO may arise from novel alternative CLN3 transcripts that have yet to be described.
4.4 Pathogenicity of the c.175G>A variant: Cellular morphology and cellular function
CLN3c.175G>A/Δ1kb patient dermal fibroblasts and lymphocytes were previously found to contain large empty vacuoles (Chen et al., 2019), consistent with observations reported in other patients with non-syndromic CLN3 retinopathy (Ku et al., 2017). Previously, iPSC derived-neural progenitor cells generated from patients with JNCL were shown to accumulate lysosomal SCMAS and intracellular storage material with a mixed curvilinear/fingerprint profile (Lojewski et al., 2014; Ryazantsev et al., 2007). In the present study, PPCs in patient-derived NRO displayed vacuolization as well as accumulation and mislocalization of peroxisomes (Figure 3b). SCMAS has been shown to accumulate in intracellular inclusion bodies in cells of the central nervous system and muscles of patients with JNCL, and has been attributed to defects in lysosomal maturation and mitochondrial autophagy (Ryazantsev et al., 2007). Here, we demonstrated significantly increased levels of SCMAS in uncorrected patient (CLN3c.175G>A/Δ1kb) iPSC, CM, and NRO cultures compared with gene-corrected (CLN3WT/Δ1kb) controls. Together, these results demonstrate the presence of pathological changes consistent with CLN3 deficiency in patient-derived cells. Correction of the c.175G>A mutation was sufficient to prevent these pathological changes from emerging, providing direct evidence supporting the pathogenicity of this variant in these cell types. Although a gene editing clinical trial for inherited retinal disease is currently underway (NCT#03872479), there remain unanswered questions regarding the optimal method for delivery of guides, Cas9 and repair templates to the affected retinal cells; the efficiencies of homology directed repair (compared with high cutting efficiencies of Cas9); and the possibility of mutagenic off-target effects. Nevertheless, since it is estimated that gene correction in ≥10% of photoreceptor cells could lead to meaningful clinical improvements for some IRDs (Stefanidakis et al., 2018), continued improvements to clinical gene editing techniques may lead to new therapeutic approaches for CLN3 disease.
4.5 Conclusions
In summary, we demonstrated the expression of novel CLN3 transcript variants in adult human retina and iPSC-derived NRO. Patient iPSC-derived cells displayed aberrant CLN3 splicing and protein expression as well as hallmarks of CLN3 deficiency, including vacuolization, increased lysosomal content, and SCMAS accumulation. These defects were prevented by correction of the c.175G>A variant, demonstrating the pathogenicity of the mutation and providing proof of principle for the therapeutic benefit of treating one variant in this recessive disease. Future studies aimed at unraveling the complexities of retinal- and neural-specific CLN3 splicing and protein translation are needed to achieve a better understanding of the roles of the different tissue and developmental isoforms. This knowledge will provide direction for developing neuroprotective therapies for syndromic and non-syndromic CLN3 disease.
ACKNOWLEDGMENTS
The authors acknowledge the generous donations from the Miocevich family, Saleeba family, McCusker family, Australian Foundation for the Prevention of Blindness and the Australian International Postgraduate PhD Scholarship (awarded to X.Z.).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
FKC, SM, and XZ conceived the project. FKC, SM, JAT, TLM, TML, and JNDR obtained funding for the project. XZ, DZ, SCC, ZH, and LJ performed laboratory work. XZ, SM, JAT, and FKC performed data analysis. XZ and SM wrote the manuscript. FKC, JAT, TLM, TML, and JDR performed clinical analysis and genetic diagnosis of the patient. All authors reviewed and approved the manuscript.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




