Different prevalence of T2DM risk alleles in Roma population in comparison with the majority Czech population
Abstract
Background
The Czech governmental study suggests up to a 25% higher prevalence of type 2 diabetes mellitus (T2DM) in the Roma population than within the majority population. It is not known whether and to what extent these differences have a genetic background.
Methods
To analyze whether the frequencies of the alleles/genotypes of the FTO, TCF7L2, CDKN2A/2B, MAEA, TLE4, IGF2BP2, ARAP1, and KCNJ11 genes differ between the two major ethnic groups in the Czech Republic, we examined them in DNA samples from 302 Roma individuals and 298 Czech individuals.
Results
Compared to the majority population, Roma are more likely to carry risk alleles in the FTO (26% vs. 16% GG homozygotes, p < .01), IGF2BP2 (22% vs. 10% TT homozygotes, p < .0001), ARAP1 (98% vs. 95% of A allele carriers, p < .005), and CDKN2A/2B (81% vs. 66% of TT homozygotes, p < .001) genes; however, less frequently they are carriers of the TCF7L2 risk allele (34% vs. 48% of the T allele p < .0005). Finally, we found significant accumulation of T2DM-associated alleles between the Roma population in comparison with the majority population (25.4% vs. 15.2% of the carriers of at least 12 risk alleles; p < .0001).
Conclusion
The increased prevalence of T2DM in the Roma population may have a background in different frequencies of the risk alleles of genes associated with T2DM development.
1 INTRODUCTION
Type 2 diabetes mellitus (T2DM, previously known as insulin-independent diabetes) is a common civilization disease that affects the metabolism of glucose. T2DM is characterized by insulin deficiency, which is caused by pancreatic β-cell dysfunction and insulin resistance in organs. T2DM patients have increased blood glucose concentrations, and they suffer from many microvascular complications, such as diabetic nephropathy, neuropathy, and retinopathy (Zheng, Ley, & Hu, 2018). Diabetes is one of the major risk factors for cardiovascular diseases; among patients with myocardial infarction or stroke, there is a significantly higher prevalence of T2DM in comparison to the general population (Bhupathiraju & Hu, 2016; Hubacek et al., 2017; Peters, Huxley, & Woodward, 2014).
The incidence of T2DM in industrial countries is estimated to be approximately 8% in Caucasians (Emerging Risk Factors Collaboration, 2010), but it significantly differs between different ethnic groups. The highest prevalence has been described in populations surrounding the Persian Gulf, reaching almost 30% (Alhyas, McKay, & Majeed, 2012). Our awareness of the prevalence of T2DM in the Roma minorities is sparse. In fact, there are just a few studies focused on this topic (reviewed by Nunes, Kučerová, Lukáč, Kvapil, & Brož, 2018; Vozarova de Courten et al., 2003); unfortunately, they do not reach the representative sample standards and are low in number. Nevertheless, they suggest that the prevalence of diabetes in Roma increased in comparison with the general majority populations. In agreement with these observations, in the Czech Republic, the governmental study suggests up to a 25% higher prevalence of T2DM in the Roma minority than in the majority population (http://www.mzcr.cz/verejne/dokumenty/zprava-o-zdravi-obyvatel-ceske-republiky2014-_9420_3016_5.html [document in Czech, accessed November 2019]).
The pathogenesis of T2DM development is multifactorial with both genetic and environmental (agricultural policies, physical activity, sleep, food availability, and environment) factors (Bhupathiraju & Hu, 2016). It is not known whether and to what extent the interethnic differences in T2DM prevalence reflect the different genetic backgrounds between the examined populations and to what extent unhealthy lifestyle could be responsible.
Genome-wide association studies detected dozens of single nucleotide polymorphisms (SNPs) within the different genes that are associated with an increased risk of T2DM (Kodama et al., 2018; McCarthy, 2017). Talmud et al. (2015) summarized a list of 65 SNPs and created a gene score that could be informative for the estimation of increased T2DM risk.
Based on the genome-wide association studies (GWAS) results and especially on the published T2DM-associated gene score (Kodama et al., 2018; McCarthy, 2017; Talmud et al., 2015), we selected SNPs within the genes for FTO (OMIM acc. No. 610966; nucleic acid demethylase; rs17817449), TCF7L2 (OMIM acc. No. 602228: transcription factor implicated in blood glucose homeostasis; rs7903146), CDKN2A/2B (OMIM acc. No. 60060 and 600431; cyclin-dependent kinase inhibitors; rs10811661), MAEA (OMIM acc. No: 606801; erythroblast macrophage protein, homologue of the yeast glucose-induced degradation-deficient protein; rs6819243), TLE4 (OMIM acc. No: 605132; complex locus of several transcription factors; rs17791513), IGF2BP2 (OMIM acc. No: 608289; insulin-like growth factor 2 mRNA-binding protein 2; rs4402960), ARAP1 (alias CENTAURIN, OMIM acc. No: 606646; possibly involved in cell signaling and communication; rs1552224), and KCNJ11 (OMIM acc. No: 600358; kidney potassium channel; rs5215), which are considered to be the most powerful genetic determinants of T2DM development in Caucasians.
Our study was focused on the analysis of the differences in the frequencies of genotypes of the FTO, TCF7L2, CDKN2A/2B, MAEA, TLE4, IGF2BP2, ARAP1, and KCNJ11 genes between two major ethnic groups living in the Czech Republic. We tested the theory that there are significant differences in genotype frequencies of these genes between the Roma minority inhabiting the region of the Czech Republic and the majority of the Czech population.
2 MATERIAL AND METHODS
2.1 Ethical compliance
All subjects involved in the study provided written informed consent. The study protocol was approved by the institutional ethics committee and conducted according to the Good Clinical Practice guidelines and in agreement with the Helsinki Declaration of 1975.
2.2 Study populations
We examined two ethnically distinct populations inhabiting an identical region of South Bohemia (Table 1). Six hundred unrelated adults (at least 18 years old at the time of examination) were included in the study (Adámková et al., 2015; Hubacek et al., 2018; Šedová et al., 2015). Czech Roma (N = 302) were recruited using snowball sampling (Hughes, Fenton, Hine, Pilgrim, & Tibbs, 1995) and Czech Caucasians/Slavs (N = 298) were recruited using quota sampling (for more details, see Adámková et al., 2015; Šedová et al., 2015; Walter, 1989). Ethnicity was based on self-reported information.
| Roma | Majority slavs | |
|---|---|---|
| N | 302 | 298 |
| Age (y) | 39.2 ± 12.8 | 39.5 ± 15.1 |
| % of females | 50 | 50 |
| BMI (kg/m2) | 29,9 ± 5,6 | 25.0 ± 6,0 |
| SBP (mm Hg) | 124,3 ± 20,9 | 124.9 ± 14.4 |
| DBP (mm Hg) | 76,6 ± 12,71 | 77.2 ± 10.6 |
| Total cholesterol (mmol/L) | 5.1 ± 1.4 | 5.1 ± 1.1 |
| Glycemia 6 mmol/L and more (%) | 58.6 | 23.3 |
| Total body fat 25% and more (%) | 69.4 | 61.7 |
| % of alcohol consumersa | 10.5 | 16.4 |
- a Defined as a consumption the day before the examination
2.3 DNA analyses
DNA was isolated using the “Xtreme DNA Isolation Kit” from buccal cells obtained through “DNA buccal swabs” (both Isohelix, Cell Projects Ltd, UK) according to conditions specified by the manufacturer.
Genotypes of interest were analyzed using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method (polymorphisms within the FTO (Hubáček et al., 2015), TCF7L2, CDKN2A/2B, MAEA, IGF2BP2, and TLE4 or by TaqMan assays (KCNJ11 and ARAP1 variants). Restriction fragments were separated using a 10% polyacrylamide gel. Fermentas International Inc. (Burlington, Ontario, Canada) provided all PCR chemicals, and PCRs were performed on the MJ Research DYAD Disciple PCR device.
Details about the oligonucleotides and restriction enzymes were used, PCR conditions or TaqMan assay ID numbers are summarized in Table 2.
| Polymorphism |
Primer sequences or Taqman assay ID |
PCR product | Enzyme | Size of restriction fragments (bp) | Allele |
Effect size per RA9−11,30 |
|---|---|---|---|---|---|---|
| FTO rs17817449 |
5′ ggtgaagaggaggagattgtgtaactgg 5′ gaagccctgagaagtttagagtaaattggg |
198 bp | AlwNI |
198 99 + 99 |
G T |
~1.2–1.5 |
|
TCF7L2 rs7903146 |
5′ gaacaattagagagctaagcactttttaggta 5′ tgtccagggcccctctaacctt |
155 bp | Rasi |
155 123 + 32 |
T C |
~1.3–2.0 |
|
CDKN2A/2B rs10811661 |
5′ tgaagacattagaacaccataacctttcc 5′ taggaggagccagaagacagatcagg |
143 bp | BspHI |
143 94 + 49 |
C T |
~ 1.2–1.5 |
|
MAEA rs6819243 |
5′ aaccttcaggttggccttgatgcctc 5′ ttcatccccaggtcaggaaactcc |
259 bp | MvaI |
189 + 70 95 + 94 + 70 |
T C |
~1.2 |
|
IGF2BP2 rs4402960 |
5′ aagcaggtttggaaccctggaattcc 5′ tctagcctgggcgacagagcaagactcc |
220 bp | MseI |
220 111 + 109 |
G T |
~1.2 |
|
TLE4 rs17791513 |
5′ aagagagtgctgaataggaaccatttgtcc 5′ taatgcttcagactatatccccttatcga |
136 bp | Bsu15I |
136 107 + 29 |
G A |
~1.2–1.5 |
|
KCNJ11 rs5215 |
C_2991148_10 | n.a. | n.a. | n.a. |
C T |
~1.15 |
|
ARAP1 rs1552224 |
C_1953903_10 |
n.a. | n.a. | n.a. |
A C |
~1.15 |
Note
- T2DM-associated allele is in bold. RA – risky allele; rs17817449 – NC_000016.10:g.53779455T>G; rs7903146 – NC_000010.11:g.112998590C>T; rs10811661 – NC_000009.12:g.22134095T>C; rs6819243 – NC_000004.12:g.1299457T>C; rs4402960 – NC_000003.12:g.185793899G>T, rs17791513 – NC_000009.12:g.79290675A>G; rs5215 – NC_000011.10:g.17387083C>T; rs1552224 – NC_000011.10:g.72722053A>C.
2.4 Statistical analysis
The deviance of genotype frequencies among the groups was analyzed according to Hardy-Weinberg equilibrium (www.tufts.edu/~mcourt01/Documents/Court%20lab%20-%20HW%20calculator.xls). Differences in allelic and genotype frequencies were compared using an online chi-square test (www.socsistatistics.com). Comparisons were performed in “AA vs. Aa vs. aa” model. In the cases where fewer than five subjects were present in some genotype category in at least one group, the rare homozygotes were pooled with heterozygotes. To calculate the unweighted gene score, the number of risk alleles [risk category was based on the publication of Talmud et al. (2015)] was summarized for each subject, and the values were compared between the ethnicities using a chi-square test. For this comparison, subjects with more than one missing genotype were excluded, leaving 287 subjects (96.3%) in the non-Roma majority Slavs and 295 (97.7%) subjects in the Roma minority population. The missing genotypes were artificially imputed as described in detail in Hubacek et al. (2019) Altogether, 48 genotyping results (1.05%) were imputed, 26 in the majority (1.13%), and 22 in Roma subjects (0.93%). In the majority, most of the imputations (N = 12) were necessary in the CDKN2A/2B gene (rs10811661), and in Roma subjects, 11 imputations were necessary for the TCF7L2 (rs7903146) gene.
As there were very low numbers of subjects with 6 and 7 risk alleles, they comprised, together with carriers of 8 risk alleles, the “low-risk” allele subgroup. In contrast, subjects with 12–16 alleles comprised the “high-risk” allele subgroup. The mean and S.D. of risk alleles were calculated for each ethnic group and compared by the two-tailed t-test.
Due to the large number of comparisons, a p < 0.01 was considered significant.
2.5 Data availability
The data that support the findings of this study are available on request from the corresponding author at [email protected]. The data are not publicly available due to privacy and ethical reason.
3 RESULTS
The achieved call rates for individual SNPs were between 92.9% and 100% in the Czech majority population and between 93.7% and 99.4% in the Roma population. Within the individual ethnic groups, no significant gender differences in genotype frequencies were observed (results not presented in detail).
3.1 Individual SNPS
The frequency of genotypes of most analyzed variants differed significantly between the examined ethnicities (for more details see Table 3). Compared to the majority population, Roma were more likely to carry risk alleles/genotypes of the FTO (26% vs. 16% GG homozygotes, p < .01), CDKN2A/2B (81% vs. 66% TT homozygotes, p < .001), IGF2BP2 (22% vs. 10% TT homozygotes, p < .0001), and ARAP1 (98% vs. 95% A allele carriers, p < .005) genes.
| Czech majority | Roma minority | p | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | % | CR | HWE | N | % | CR | HWE | |||
| FTO | ||||||||||
| rs17817449 | ||||||||||
| GG | 47 | 15.8 | 76 | 25.3 | ||||||
| GT | 161 | 54.0 | 100.0 | 0.08 | 154 | 51.3 | 99.4 | 0.64 | .009 | |
| TT | 90 | 30.2 | 70 | 23.3 | ||||||
| TCF7L2 | ||||||||||
| rs7903146 | ||||||||||
| CC | 151 | 51.9 | 186 | 65.7 | ||||||
| CT | 120 | 41.3 | 97.7 | 0.56 | 93 | 32.9 | 93.7 | 0.05 | .0008 | |
| TT | 20 | 6.9 | 4 | 1.4 | ||||||
| CDKN2A/2B | ||||||||||
| rs10811661 | ||||||||||
| TT | 188 | 67.9 | 235 | 81.3 | ||||||
| TC | 79 | 28.5 | 92.9 | 0.64 | 51 | 17.6 | 95.7 | 0.90 | .0002 | |
| CC | 10 | 3.6 | 3 | 1.0 | ||||||
| MAEA | ||||||||||
| rs6819243 | ||||||||||
| TT | 275 | 94.5 | 273 | 92.5 | ||||||
| TC | 16 | 5.5 | 97.7 | 0.63 | 22 | 7.5 | 97.7 | 0.51 | .34 | |
| CC | 0 | 0.0 | 0 | 0.0 | ||||||
| TLE4 | ||||||||||
| rs17791513 | ||||||||||
| AA | 260 | 88.7 | 271 | 90.9 | ||||||
| AG | 32 | 10.9 | 98.3 | 0.99 | 26 | 8.7 | 98.7 | 0.67 | .38 | |
| GG | 1 | 0.4 | 1 | 0.3 | ||||||
| KCNJ11 | ||||||||||
| rs5215 | ||||||||||
| CC | 113 | 39.8 | 121 | 40.5 | ||||||
| CT | 121 | 42.6 | 95.3 | 0.08 | 131 | 43.8 | 99.0 | 0.25 | .83 | |
| TT | 50 | 17.6 | 47 | 15.7 | ||||||
| IGF2BP2 | ||||||||||
| rs4402960 | ||||||||||
| GG | 134 | 46.4 | 84 | 28.2 | ||||||
| GT | 125 | 43.3 | 97.0 | 0.91 | 149 | 50.0 | 98.7 | 0.94 | .0001 | |
| TT | 30 | 10.4 | 65 | 21.8 | ||||||
| ARAP1 | ||||||||||
| rs1552224 | ||||||||||
| AA | 204 | 70.1 | 235 | 80.2 | ||||||
| AC | 72 | 24.7 | 97.7 | 0.01 | 53 | 18.1 | 97.0 | 0.33 | .007 | |
| CC | 15 | 5.2 | 5 | 1.7 | ||||||
Note
- T2DM-associated allele is in bold. rs17817449 – NC_000016.10:g.53779455T>G; rs7903146 – NC_000010.11:g.112998590C>T; rs10811661 – NC_000009.12:g.22134095T>C; rs6819243 – NC_000004.12:g.1299457T>C; rs4402960 – NC_000003.12:g.185793899G>T, rs17791513 – NC_000009.12:g.79290675A>G; rs5215 – NC_000011.10:g.17387083C>T; rs1552224 – NC_000011.10:g.72722053A>C.
- Abbreviations: CR – call rate; HWE – Hardy–Weinberg equilibrium.
In contrast, in the Roma population, there were fewer carriers of the TCF7L2 risk allele (34% vs. 48% of the T allele p < .0005) in comparison with the majority population.
Finally, we did not detect differences between the Czech majority population and Roma living in the Czech Republic for the allelic/genotype frequencies of the polymorphisms within the MAEA, TLE4, and KCNJ11 genes. For the first two mentioned, the cause could be the generally low frequency (in both cases below 6% in both ethnicities) of the minor allele, suggesting the possible insufficient power of the study.
3.2 Gene score
The cumulative prevalence of the risk alleles within both examined groups is summarized in Table 4 and Figure 1. The span of the numbers of risk alleles was wide. The minimum observed was, however, relatively high. Everybody was a carrier of at least 6 risk alleles, and one subject was carrier of all possible 16 risk alleles. The unweighted gene score values were between 6 and 16 in the Czech majority population and between 7 and 15 in the Roma population. Means of risk alleles differed significantly (p < .00005) between the two groups. There were 9.97 ± 2.28 risk alleles per person in the majority and 10.52 ± 2.25 risk alleles per person within the Roma subjects.
| N | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | |
| Majority | 8 | 2.8 | 11 | 3.8 | 34 | 11.8 | 53 | 18.5 | 69 | 24.0 | 68 | 23.7 | 30 | 10.5 | 9 | 3.1 | 3 | 1.0 | 1 | 0.3 | 1 | 0.3 |
| Minority | 0 | 0.0 | 7 | 2.4 | 13 | 4.4 | 52 | 17.6 | 83 | 28.1 | 65 | 22.0 | 49 | 16.6 | 16 | 5.4 | 9 | 3.1 | 1 | 0.3 | 0 | 0.0 |
- Abbreviation: N, number of risk alleles.
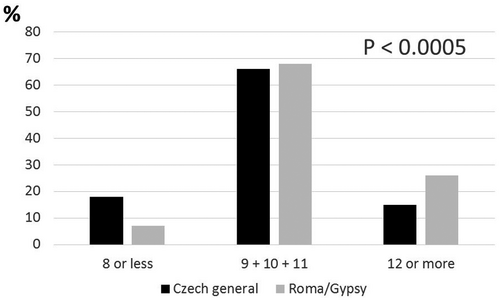
There was a significant difference (p < .0001) in the unweighted gene score value distribution between the Czech majority population and the Roma population in predefined subgroups (Figure 1).
Within the low-risk group range (8 risk alleles maximum), there were 18.4% of majority subjects and 6.6% of Roma subjects only. In contrast, within the high-risk group category (at least 12 risk alleles), 15.2% were the majority, and 25.4% were Roma subjects.
4 DISCUSSION
The results of our study support the theory that the increased prevalence of T2DM in the Roma population may be associated with different frequencies of the risk alleles of some genes associated with T2DM development.
Our study is in contrast with a recently published study focused on similar topics in the Hungarian majority and Roma populations (Werissa et al., 2019). They found slightly increased accumulation of T2DM-associated alleles within the majority population. This discrepancy with our study, however, could be based simply on the selection of genes/variants. The Hungarian authors selected 16 SNPs, but only two of them (within the TCF7L2 and CDKN2A/2B genes) were identical to our study. Interestingly, the frequencies of SNPs within these genes differ to some extent in our study and the Hungarian study. For the general Czech and Hungarian population, the minor TCF7L2 allele frequencies are highly similar (27.5% vs. 29.5%), but Hungarian Roma have a higher prevalence of the risk allele (24.2% vs. 17.8%) than Roma inhabiting the Czech region. For CDK2A/2B, the majority populations had frequencies of the risk allele of 82.8% and 82.1%, and Czech Roma were carriers of this allele (90.1%) slightly more often than Hungarian Roma (85.3). Even these incomplete comparisons suggest that there could be increased genetic heterogeneity between different Roma subpopulations across Europe. Another important issue that needs to be mentioned is the fact that a previous study (Nagy et al., 2011) suggests the Asian origin of Hungarians; thus, the genetic background of different ethnicities in the Hungarian geographic region could be historically more similar than that in other European regions.
Of the analyzed SNPs/genes, FTO is probably the most commonly studied. Minor alleles are associated with an increased risk of T2DM as well as increased risk of T2DM complications (Gaulton, 2017; Hubacek et al., 2018). FTO variants are also discussed as potential predictors of obesity treatment (Xiang et al., 2016; Zlatohlavek et al., 2013). FTO polymorphisms are the only SNPs whose frequencies were analyzed previously in more Roma populations from different regions. The prevalence of minor allele homozygotes of the rs9939609 variant (which is in non-Roma Caucasians in almost complete LD with rs17817449 variant) was 22% in the Slovak-Roma population (Mačeková et al., 2012) and, as expected, significantly associated with increased BMI values. A similar association between increased BMI and the FTO rs9939609 variant was observed in the Hungarian Roma (Nagy, Fiatal, Sándor, & Ádány, 2017) (where the prevalence of the FTO risk allele is higher) but not in the Spanish Roma population (Poveda, Ibáñez, & Rebato, 2014). These results suggest a relatively high FTO polymorphism heterogeneity effect between the Roma populations (or possibly, not such strong LD between rs9939609 and rs17817449 polymorphisms) in comparison with the European majority populations.
The TCF7L2 gene seems to be the strongest genetic predictor of T2DM development (Adams & Vella, 2018). T2DM-associated alleles of this gene are associated with reduced β-cell function (reviewed by Adams & Vella, 2018). Most interestingly, in this study, we found a higher frequency of the C allele within the Roma population. This allele is described as protective against T2DM development, and this had been confirmed also in the Czech population (Včelák et al., 2012).
The importance of CDKN2A/2B in T2DM pathology is not clear, albeit rs10811661 was identified in the first GWAS presented among the most powerful signals (Cauchi et al., 2008). However, there seems not to be an association between this variant and an increased risk of T2DM in the Czech population (Hubáček, Neškudla, Klementová, Adámková, & Pelikánová, 2013) and similarly controversial results have also been published for the Arab population (Nemr et al., 2012). In contrast, an association between this polymorphism and T2DM was found in an Asian Indian (Chidambaram et al., 2016) and a Pakistani populations (Rees et al., 2011), which are populations from the geographical region where the European Roma seem to originate.
Most of the studies of IGF2BP2 are focused on gestational diabetes, but a global meta-analysis confers an association with T2DM both for Caucasians and for Indian populations (Zhao et al., 2012).
For the ARAP1 (alias CENTD2) variant, the vast majority (>95%) of subjects in both ethnicities are carriers of at least one risk allele here. This makes the utility of this variant questionable for T2DM estimation at the population level. The major allele is associated with reduced insulin release (Nielsen et al., 2011) and was associated with protection from T2DM in northern-Indian women using the WHO 2013 (but not using the WHO 1999) criteria (Arora et al., 2018).
The T2DM-associated risk potential of only four genes of our set [for FTO (Hubacek et al., 2018), IGF2BP2, (Gu et al., 2012), CDKN2A/2B (Hubáček et al., 2013), and TCF7L2 (Včelák et al., 2012)] was examined in the Czech majority population. Thus, although we are aware of the potential inaccuracy, for creation of the unweighted gene score, we have used the risk status based on the study of Talmud et al. (2015). The use of aggregated risk score, summarizing the effects of more genes in one single value, should improve the predictive ability of genetic testing. Here, we clearly show that Roma subjects are carriers of more T2DM-associated alleles.
Our study is not the only one that has described the differences in genetic background between the Roma and majority Caucasian populations.
For example, in Roma, allelic differences in lactate dehydrogenase (Hubácek et al., 2017b), drug metabolism enzymes (Tomas et al., 2017, Dlouhá et al., in press), or osteoprotegerin (Mydlárová Blaščáková et al., 2017) polymorphisms clearly underlie genetic differences that could have important clinical (optimal treatment or dietary recommendations) or live expectancy consequences.
Previous studies and our results show that the Roma are genetically distinct from both the European majority population and the original Asian populations, where the origin of European Roma gypsies is dated. The differences could be due to random genetic drift, mostly the founder effects rather than unlikely selection pressure or environment adaptation, as these factors were identical in past centuries for majority and minority populations inhabiting the same European region.
We are aware of the limitations of our study. The major one is that for the gene score calculation, we had to use the results obtained for other European populations. Nevertheless, it cannot be excluded that (especially in the case of the Roma minority) the associations between the analyzed genes and T2DM could differ from the expected directions. We have not confirmed the association with T2DM for the CDKN2A/2B rs10811661 SNP (Hubáček et al., 2013). The status between the risk of T2DM in the Czech population is not known for the rest of the analyzed genes; thus, the potential limitation of the present study is that we used allelic gene score based on results obtained from other population (Talmud et al., 2015) and we decide not to use the weighted gene score which takes into account also the effect size of individual gene variants. This, however, does not reduce the importance of our findings; in fact, quite the contrary. If in the future the genetic risk score will be used for risk estimation, the analysis needs to be performed in young asymptomatic subjects, where the weighted gene values would be definitely different from the values obtained for the population, where increased nongenetic risk factors are presented.
The genetic tools, likely the population-specific genetic risk scores, in the future could improve the in-time identification of the high-risk subsets of the population. Accumulating evidence suggests that genetic testing will have an irreplaceable role in personalized therapy and clinical decision making (Rodríguez Vicente, Herrero Cervera, Bernal, Rojas, & Peiró, 2018). To achieve this goal, examinations of a higher number of genetic polymorphisms, especially in minorities, interaction analysis, and detailed population-specific gene score calculation are the further necessary steps. It is clear that there are important differences between populations and ethnicities.
We conclude that there is a significant genetic difference between the Czech majority population and the Roma population. The increased prevalence of T2DM in the Roma population could be based on an increased cumulative number of T2DM-associated common alleles within different genes.
ACKNOWLEDGEMENTS
The study was supported by Ministry of Health of the Czech Republic, grant no. NV18-01-00046, all rights reserved; projects number LD14114 implemented under the financial support of the Ministry of Education, Youth and Sports within COST (Cooperation On Scientific and Technical Research), entitled “Obesity and overweight in the Romany minority in the Region of South Bohemia”, and by the project supported by Ministry of Health Czech Republic – conceptual development of research organization (“Institute for Clinical and Experimental Medicine – IKEM, IN 00023001”).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTION
JAH – Writing – Original Draft; Methodology; Investigation – genotyping; Funding acquisition; Supervision; Data analysis; Conceptualization. LŠ – Writing – Review & Editing; Investigation; Data Curation. VO – Writing – Review & Editing; Investigation; Data Curation. VA – Writing – Review & Editing; Methodology; Investigation; Resources; Supervision; Conceptualization. VT – Writing – Review & Editing; Funding acquisition; Methodology; Investigation; Resources; Project administration; Visualization; Formal analysis; Conceptualization.