Association of polymorphism in heat shock protein 70 genes with type 2 diabetes in Bangladeshi population
Funding information
The work has been supported by the fund of Special allocation of Ministry of Science and technology, Government of the people's Republic of Bangladesh in the fiscal year of 2015- 16. The project ID was MEDI; S- 250.
Abstract
Background
Type 2 diabetes mellitus (T2DM) is a chronic disorder of which stress is a major contributor. Under stressful condition, body synthesizes a family of molecular chaperone called Heat-shock proteins (HSPs). Current study assessed the frequency and association of HSP70-hom + 2,437 T/C polymorphism with T2DM risk among Bangladeshis.
Methods
This polymorphism was selected through bioinformatics analyses and identified by PCR-RFLP method.
Results
Bioinformatics analysis identified this SNP as missense mutation which could destabilize the final HSP product. Heterozygous mutant (CT) genotype was significantly associated with T2DM incidence among the studied populations (p = .015). Further analysis revealed a strong association with female patients (p = .002), while the male group showed no association (p = .958). Moreover, the C allele was significantly associated among all diabetic patients (p = .016) and particularly in the female patient group (p = .001). However, under stressful condition, males with CT genotype were at high risk for T2DM incidence whereas, females with CT genotype showed no significant association.
Conclusions
HSP70-hom + 2,437 T/C polymorphism was found to be significantly associated with T2DM incidence in the Bangladeshi population in both stress-dependent and independent manners.
1 INTRODUCTION
Type 2 diabetes mellitus (T2DM) is an ever-increasing chronic metabolic disorder with a strong genetic predisposition. Globally, 1 of 11 adults bearing diabetes while it is forecasted to be 1 in 10 by the year 2040 (IDF Atlas, 2015). The high incidence of diabetes creates a major problem in developing countries like Bangladesh where the prevalence of diabetes are estimated to elevate from 9.2% to 13% by 2030 (IDF Atlas, 2015; Rahman et al., 2015).
Stress is one of the major contributors to diabetes. Oxidative stress has been proposed as the root cause underlying the development of insulin resistance, β-cell dysfunction, impaired glucose tolerance, and T2DM through the production of reactive oxygen species. It has also been implicated in the progression of long-term diabetes-associated complications (S. J. Kelly & Ismail, 2015; Pouwer, Kupper, & Adriaanse, 2010). Stresses such as heat shock, ischemia, and other types of cellular stresses induce synthesis of one of the most ubiquitous and highly conserved molecular chaperone, heat shock proteins (HSPs) (Gehrmann et al., 2005; Lindquist, 1986). The 70-kDa HSP (HSP70) family is the most abundant in eukaryotic cells and is essential for cell survival under stressful conditions (Kelly, 2002). In humans, there are three main genes (HSPA1A, HSPA1B, and HSPA1L) of HSP70 family and the coding proteins are defined as HSP70-1, HSP70-2, and HSP70-hom, respectively. Through their effect on insulin sensitivity, HSPs have also been reported to be involved in diabetes (Macario & De Macario, 2000).
HSP70 gene polymorphism is reported to be associated with an increased risk of developing various disorders including T2DM (Chouchane et al., 2001; Ghanayem et al., 2014; He, Deng, & Luo, 2014; Pociot, Rønningen, & Nerup, 1993; Umapathy, Krishnamoorthy, & Muthukumaran, 2012; Umapathy et al., 2012) . One of the most studied single-nucleotide polymorphisms (SNPs) of HSPA1L is located at position + 2437 of HSP70-hom (rs2227956). This is a nonsynonymous missense mutation observed in the coding region of HSP70-hom leading to the change in amino acid from threonine to methionine that affects the stability and activity of this protein (Singh et al., 2006). Moreover, this SNP could affect HSP70 expression or function and further contribute to disease susceptibility and stress tolerance (Favatier, Bornman, Hightower, Günther, & Polla, 1997). Several epidemiological studies have been done to deduce the relationship of this polymorphism with obesity, DM and Diabetes nephropathy (DN) (Ghanayem et al., 2014). These studies revealed both significant and nonsignificant association between HSP70-hom + 2437 T/C polymorphism and T2DM (Ghanayem et al., 2014; Umapathy et al., 2012). The SNP, HSP70-hom + 2437 T/C was picked up for this study based on bioinformatics analysis which was authenticated by previous association studies. The soundness of this particular SNP study was confirmed through the measurement of its deleterious effect (Mooney, 2005) on the stabilization of the protein using different bioinformatics tools. We have further investigated the frequency of this polymorphism and its association with T2DM in the Bangladeshi mainstream population. To our knowledge, this is the first genetic association study on this SNP ever conducted on Bangladeshi patients with diagnosed T2DM cases. Also, we investigated the cumulative effect of stress and polymorphism on T2DM prevalence in the studied population.
2 MATERIAL AND METHODS
2.1 Ethical compliance
The case–control study approved by the ethics committee on National Institute of Biotechnology (NIB). The approval ID is NIBREC2015-02.
2.2 Selection and in-silico validation of SNP HSP70-hom +2437 (T/C)
The information on target SNP (rs2227956) was derived from Online Mendelian Inheritance in Man (OMIM) and NCBI dbSNP (Database of Single-Nucleotide Polymorphism) (Hamosh, Scott, Amberger, Bocchini, & McKusick, 2005; Sherry et al., 2001). Protein sequence and its structure were retrieved by using Uniprot (Universal Protein Resource) and Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB), respectively (Consortium, 2016; Rose et al., 2010). A web server called ConSurf was utilized to predict evolutionary conserved regions of the HSPA1L protein. A program called MODELLER 9v11 (Šali, Potterton, Yuan, van Vlijmen, & Karplus, 1995) through HHpred (Söding, Biegert, & Lupas, 2005) was used to determine the 3D structure of Wild-type and Mutant Protein encoded by HSPA1L gene. To find out the structure of the closest-related proteins, we submitted the protein sequence to HHPred Modeller tool and performed BLAST against the Protein Database (PDB). The best three-dimensional models were selected using ProCheck (Laskowski, MacArthur, Moss, & Thornton, 1993) and Verify3D (Lüthy, Bowie, & Eisenberg, 1992). Another tool named Yet Another Scientific Artificial Reality Application (YASARA) was introduced to calculate the energy minimization of both wild and possible mutant-type protein models (Kuszewski, Gronenborn, & Clore, 1997). This program yields the energy (START vs. END) that ensures accuracy of designed molecules. To confirm the variability in 3D structure of the proteins, an extensive virtual screening was performed with a suitable drug. At the same time, the drug's chemical structure, molecular weight, generic name, and binding capacity were also confirmed by bioinformatics analysis.
2.3 Study population
The case–control study comprised of 216 clinically confirmed T2DM patients (mean age 50.38 ± 12.35 years) using glucose oxidase-peroxidase (GOD-POD) method and oral glucose tolerance test (OGTT) according to the WHO guideline from Diabetes and health awareness center, Kustia,, Bangladesh and Medical Centre of National Institute of Biotechnology, Dhaka, Bangladesh and 126 healthy controls (mean age 44.58 ± 12.4 years) from the general Bangladeshi population. The control respondent was further confirmed by measuring fasting plasma glucose (via the glucose oxidase-peroxidase [GOD-POD] method) (Meites & Saniel-Banrey, 1973). Informed written consent was taken from all subjects enrolled in the study in accordance with the Helsinki declaration. All participants also completed a questionnaire and participated in an interview. The administered questionnaire included information about age, sex, BMI, food habit, physical exercise, and history of first degree relatives, etc. As all patients included in this study were prediagnosed and most of the demographic parameters were in control, the information (except age and sex) were not included in this study.
2.4 Sample size calculation and power of study
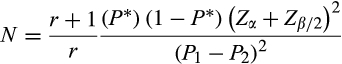
2.5 Categorization of stress
The respondents were administered a detailed questionnaire comprised of information about life event (close relative's death, accident, failure in examination etc.), mental problem/depression, work stress, worries about future planning, financial stress, and daily life stress (Knol et al., 2006; Kumari, Head, & Marmot, 2004; Norberg et al., 2007; Rod, Grønbaek, Schnohr, Prescott, & Kristensen, 2009; Toshihiro, 2008). Numerical score had been given to every stress to assess the level of stress the participants suffered. Following this, the stress score was categorized into nonstress (0–1 point) and stress (2–6 points) groups. Further, the stress group was divided into low (2–3 points) and high (4–6 points) stress according to the modified method of Rod NH et al (Rod et al., 2009).
2.6 Blood sampling and DNA extraction
Three milliliter peripheral blood was drawn in EDTA containing vacutainer (BD, Oxford, UK) Tube. Erythrocytes of the samples were lysed by osmotic shock using 20 mM Tris-HCl (pH 8.0). DNA was extracted using Genomic DNA isolation kit (FavorPrep, Favorgen, Taiwan). The DNA concentration was determined using a Nanodrop spectrophotometer (Thermo Scientific, USA). Acceptable DNA samples (A260/A280 of 1.8–2.0) were diluted to 50 ng/μl and stored at −80°C until used.
2.7 Genotyping
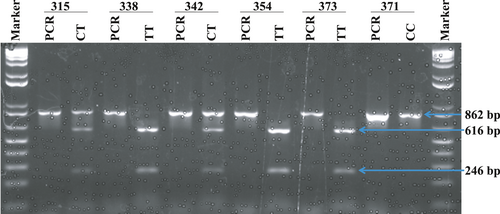
Genotype of HSP70-hom (+2437 T/C) was determined by the polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) method using the following primers 5′GGACAAGTCTGAGAAGGTACAG-3′ (forward) and 5′-GTAACTTAGATTCAGGTCTGG-3′ (reverse). Briefly, the PCR mixture contained a 1x reaction buffer (Thermo Fisher Scientific, Waltham, MA USA), 2.0 mM MgCl2, 0.2 mM dNTP, 20 picomoles of the two primers, 50–100 ng of genomic DNA and 1.125 unit of Taq DNA polymerase (Thermo Fisher Scientific, Waltham, MA USA) in a total volume of 25 μl. Amplification was accomplished by following the thermal cycling condition: 95°C for 5 min (one cycle); 95°C for 1 min, 61°C for 1 min, and 72°C for 1 min (35 cycles); 72°C for 7 min (one cycle), and hold at 4°C. The amplification product (862 bp) was visualized and documented using Gel documentation (proteinsimple, Santa Cara, CA USA) after resolving the product along with DNA size markers (1 Kb + DNA ladder, Invitrogen, USA) in 1% agarose gel and staining the gel with ethidium bromide. The resulting amplicons were subjected to restriction digestion using restriction endonuclease NcoI (New England Biolabs, UK) at 37°C for 2 hrs and then resolved in 1.5% gel.
2.8 Statistical Analysis
Different statistical analyses using SPSS (SPSS v20, IBM) and MedCalc (Medcalc, Mariakerke, Belgium) softwares were performed to validate the results generated from the study. A robust test estimating χ2 value at the 0.05% level of significance was carried out to check whether the genotypic frequencies of the polymorphism in HSPA1L deviated from Hardy–Weinberg equilibrium. The value was considered as rejected if it crossed the standard mark (χ2 > 3.84). Odds ratio (OR) with 95% confidence interval (CI) was calculated for the determination of the association of genotype and allele type with T2DM. Risk analysis by relative risk (RR) estimation with 95% CI confirmed the risk of disease occurrence for having specific genotypes. The above results having a p value ≤ 0.05 were accepted as statistically significant.
3 RESULTS
3.1 In -silico identification and validation of HSP70-hom +2437 (T/C) as a potent T2DM marker
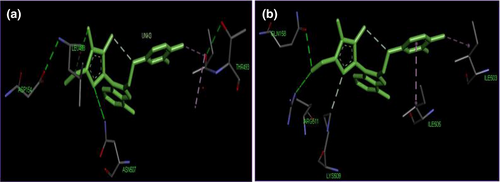
Out of 1,192 SNPs found in HSPA1L gene, 359 are missense. The target SNP (rs2227956) is one of these missense SNPs. The protein encoded by HSPA1L gene consists of 641 amino acids in which amino acid at 493 position changes from threonine (T) to methionine (M) due to this SNP. Amino acid conservation analysis by ConSurf also predicted this T amino acid as a variable one (Figure S1). We built the 3D protein structures for both wild type and possible mutant sequence (T → M) based on the PDB id 5e84_A which shows 100% identity with HSPA1L protein. The tertiary structure having a minimum energy was predicted and the quality of the model was verified through different bioinformatics tools (Figure S2, S3, S4). Our analysis revealed decreased free energy for all mutant-type proteins (−1122.1 KJ/mol) than wild types (−1111.1 KJ/mol) (Figure S5). Drug binding analysis with an experimental drug DB07045 also identified the structural variation between wild type and mutant (T493M) model (Table S1). In wild-type Thr493, Asp154, Asn507, and Leu488 were the interacted amino acid residues with the drug whereas amino acids Ile503, Ile505, Arg511, lys509, and Gln158 interacted with the mutant model (Figure 1). The interacting amino acid Thr493 is in the active site of the protein (Figure S6).
3.2 Genotypic distribution of HSP70-hom (+2437)
DNA was isolated from peripheral blood of 216 clinically confirmed diabetes patients comprised of 78 males and 138 females and 126 closely age-matched clinically confirmed healthy control subjects, including 56 males and 70 females. The good quality isolated DNA was subjected to PCR-RFLP methods. The digestion generated two fragments of 616 bp and 246 bp size upon the presence of the desired SNP (Figure 2). Genotypic distribution of HSP70-hom (+2,437) polymorphism in the studied population followed the principle of Hardy–Weinberg equilibrium (Table S2).
Genotype frequency among the male, female, and all subjects with and without type 2 diabetes was measured among the studied subjects (Table 1). Frequencies of subjects bearing the CT and CC genotypes were higher in the diabetic group than in the control when all subjects are considered. Multivariate logistic regression was done to assess risk level of genotypes and alleles of the studied SNP (Table 2). The frequency difference of CC genotype was not statistically significant (p = .725), while the CT genotype increased the risk of type 2 diabetes with a significant level (p = .015) of difference. When the frequencies of CT and CC genotypes of male and female subjects were separately compared with the corresponding control, the same scenario was found as that of all subjects. However, in the case of female subjects increased prevalence of CT genotype in patient was highly significant (p = .002). In comparison, the relation is not significant in the case of male respondents (p = .958).
| Catagories | Genotype/Allele | Control (%) | Patient (%) |
|---|---|---|---|
| All Subjects | CT | 36 (28.6) | 86 (39.8) |
| TT | 85 (67.5) | 116 (53.7) | |
| CC | 5 (3.9) | 14 (6.5) | |
| C | 46 (18.25) | 74 (26.39) | |
| T | 206 (81.75) | 318 (73.61) | |
| Male | CT | 14 (29.16) | 31 (39.74) |
| TT | 32 (66.66) | 45 (57.69) | |
| CC | 2 (4.16) | 2 (2.56) | |
| C | 18 (18.75) | 35 (22.44) | |
| T | 78 (81.25) | 121 (77.56) | |
| Female | CT | 22 (28.21) | 55 (39.85) |
| TT | 53 (67.95) | 71 (51.44) | |
| CC | 3 (3.84) | 12 (8.69) | |
| C | 28 (17.95) | 79 (28.62) | |
| T | 128 (82.05) | 197 (71.38) |
| Catagories |
Genotype/ Allele |
Unadjusted OR (95% CI); p-value | P-value | Adjusted OR (95% CI);p-valuea | P-value |
|---|---|---|---|---|---|
| All Subjects | CC | 2.08 (0.72–6.00); 0.175 | 0.031 | 1.22 (0.40–3.68); 0.725 | 0.0508 |
| CT | 1.83 (1.13–2.96); 0.015 | 1.91 (1.14–3.22); 0.015 | |||
| TT | Ref | Ref | |||
| C allele | 1.61(1.1–2.4); 0.016 | − | − | − | |
| Male | CC | 0.74 (0.10–5.57); 0.774 | 0.917 | 0.45 (0.05–3.65); 0.452 | 0.739 |
| CT | 1.10 (0.54–2.25); 0.798 | 1.02 (0.46–2.29); 0.958 | |||
| TT | Ref | Ref | |||
| C allele | 1.00 (0.54–1.7); 0.880 | − | − | − | |
| Female | CC | 2.97 (0.80–11.07); 0.105 | 0.005 | 1.96 (0.50–7.60); 0.332 | 0.009 |
| CT | 2.92 (1.47–5.81); 0.002 | 3.03 (1.48–6.20); 0.002 | |||
| TT | Ref | Ref | |||
| C allele | 2.4 (1.4–4.1); 0.001 | - | − | − |
- a Adjusted for confounding factors (Age, Sex, stress)
The comparative analysis of allele frequency between diabetic group and control subjects showed that the C allele is significantly associated among all the patients (OR = 1.61, CI: 1.1–2.4, p = .016) and frequency difference was strongly significant for female patients group (p = .001) (Table 2). However, allele frequency of C did not differ by diabetes status among male subjects (p = .880).
3.3 Association of HSP70-hom (+2437) polymorphism and stress with T2DM
The risk estimation analysis based on different combination of genotypic ratio is shown in Table S3. Higher p value made the ratio of CC/CT (OR = 1.17, p = .78) and CC/TT (OR = 2.05, p = .18) among the diabetic patients compared to the nondiabetic control group not significant, although OR and risk ratio (RR) were the highest for the CC/TT (OR = 2.05, RR = 1.28) among the three combination. However, the diabetes patient had higher ratio for CT/TT compare to the control and the result was statistically significant (OR = 1.75, p = .02; RR = 1.22, p = .02). The ratio of (CC + CT)/TT and CC/(CT + TT) among diabetes patient were substantially higher than that of control respondents, although the difference for CC/(CT + TT) was not quite statistically significant (OR = 1.68, p = .33; RR = 1.18, p = .25), but the higher ratio for (CC + CT)/TT was statistically significant (OR = 1.79, p = .01; RR = 1.23, p = .01).
Whether age and stress variables are risk factors for type 2 diabetes incidence was assessed by multivariate logistic regression (Table S4). Subjects in the age groups of (40– 60) and >60 years had 1.78× (p = .005) and 3.19× (p = .006) greater risk for type 2 diabetes respectively than group of <40 years. Overall, patients under stressful condition are more likely to develop T2DM than that of nonstressed respondent (p = .000). Moreover, when stress is divided into two groups- low stress and high stress, we found that both males (p = .000) and females (p = .000) with high stress were at high risk of diabetes mellitus, whereas the association between low stress and T2DM incidence was significant only among males (Male: p = .002; Female: p = .115). The distribution and association of the genotypes, age, and stress with T2DM have been summarized in Table 3 and Figure 3. There was no difference in T2DM incidence between CT (p = .030) and TT/CC (p = .034) genotype containing people who were in age group of 40–60 years (Table 3). In contrast, people who were more than 60 years old with CT genotype (OR = 4.636, p = .029) were more prone to T2DM than that of TT/CC genotype (OR = 3.714, p = .007) subjects (Table 3).
| Status | Coefficient | OR | 95% CI | p Value | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diabetic patients |
CT (n = 122) |
Age, ≥61 | 1.534 | 4.636 | 1.171– 18.363 | 0.029 | |||||||
| Age, 40–60 | 0.941 | 2.564 | 1.094– 6.010 | 0.030 | |||||||||
| Age, ≤39 | Reference | ||||||||||||
|
TT/CC (n = 220) |
Age, ≥61 | 1.312 | 3.714 | 1.439– 9.590 | 0.007 | ||||||||
| Age, 40–60 | 0.717 | 2.048 | 1.054– 3.978 | 0.034 | |||||||||
| Age, ≤39 | Reference | ||||||||||||
| *Corresponding control is the reference category | |||||||||||||
| Diabetic patients |
Male CT |
Stress | 2.395 | 10.971 | 2.959– 40.679 | 0.000 | |||||||
| Nonstress | Reference | ||||||||||||
|
Female CT |
Stress | 1.008 | 2.741 | 0.826– 9.091 | 0.099 | ||||||||
| Nonstress | Reference | ||||||||||||
|
All Patients CT |
Stress | 1.672 | 5.324 | 2.271– 12.480 | 0.000 | ||||||||
| Nonstress | Reference | ||||||||||||
|
Male TT/CC |
Stress | 1.051 | 2.861 | 1.128– 7.257 | 0.027 | ||||||||
| Nonstress | Reference | ||||||||||||
|
Female TT/CC |
Stress | 0.985 | 2.677 | 1.322– 5.421 | 0.006 | ||||||||
| Nonstress | Reference | ||||||||||||
|
All Patients TT/CC |
Stress | 1.01 | 2.747 | 1.575– 4.972 | 0.000 | ||||||||
| Nonstress | Reference | ||||||||||||
| *Corresponding control is the reference category | |||||||||||||
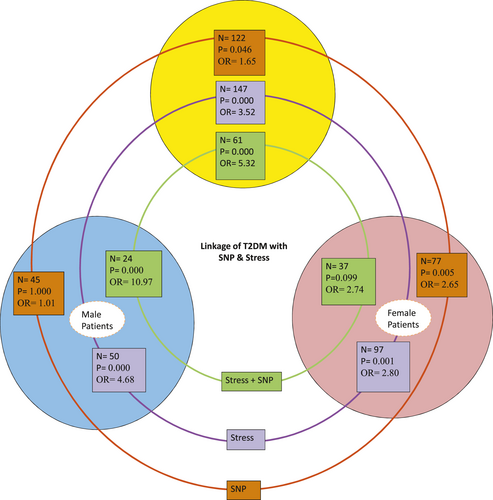
When we studied cumulative effect of the polymorphism with stress and gender on the onset of T2DM, it was found that female with CT (OR = 2.741, p = .099) or TT/CC (OR = 2.677, p = .006) genotype showed almost same level of risk, albeit relation with CT genotype was statistically insignificant. However, male with CT genotype (OR = 10.971, p = .000) were at significantly higher risk than TT/CC genotype (OR = 2.861, p = .027) bearing male. Moreover, sex independent analysis of all patients with CT genotype (OR = 5.324, p = .000) under stress was at significantly more risk to T2DM in comparison to TT/CC genotype (OR = 2.747, p = .000).
4 DISCUSSION
The rapidly increasing number of diabetic patients becomes a global burden especially for health sector in low- and middle- income countries including Bangladesh (Bleich, Koehlmoos, Rashid, Peters, & Anderson, 2011). Many reasons such as obesity, lack of physical activity, food habit, sedentary job nature and genetic makeup are factors accounting for developing diabetes (Lyssenko & Laakso, 2013; Vilchis-Gil, Galván-Portillo, Klünder-Klünder, Cruz, & Flores-Huerta, 2015). Another cause is stress, which plays important role in the etiology of T2DM (S. J. Kelly & Ismail, 2015; Pouwer et al., 2010). Type 2 diabetic patients not only have to cope with this chronic disease, they are also at increased risk for several diseases like coronary heart disease, peripheral vascular disease, retinopathy, nephropathy, and neuropathy (Pouwer et al., 2010). To fight and control T2DM we have to seek out an alternative way of diagnosis and treatment based on patient's genetic information. This requires a deep insight into the etiology of this disease including associated single nucleotide polymorphism (SNP).
Various stresses trigger secretion of cortisol and norepinephrine which in turn raise blood glucose level by stimulating hepatic gluconeogenesis and insulin resistance (Bleich et al., 2011; Knol et al., 2006; Kumari et al., 2004; Norberg et al., 2007; Rod et al., 2009; Toshihiro et al., 2008; Zhang et al., 2006). It has been shown in the current study that highly stressful life leading women experienced 1.25 × higher T2DM prognosis than that of men, whereas women were in 1.26 × lower risk to diabetic mellitus incidence than that of men who were in low level of stress (Table S4).
Age also plays a vital role in the onset of diabetes (Cowie & Eberhardt, 1995). In south-east Asia almost 97% diabetic patients are 40 years old or more (IDF Atlas, 2017). In Bangladesh, the reported age of diabetes is ≥40 years in 71% urban and 85% rural female, while in the case of male the proportion is 85.5% urban and 86.5% in rural population (IDF Atlas, 2017). The current study also pinpointed an exponential increase in the risk of onset of T2DM with the increase of age when 40 years was chosen as the reference (Table S4).
Among the genetic factors particularly SNPs alleged for diabetics, HSP70-hom + 2437T/C was targeted for the current study. However, there are contrasting association studies on this SNP (Buraczynska, Swatowski, Buraczynska, Dragan, & Ksiazek, 2009; Mir, Pugazhendhi, Paul, Nair, & Ramakrishna, 2009; Pociot et al., 1993). Therefore, before the analysis of the blood samples, we confirmed it's suitability as a candidate SNP marker for T2DM by building 3D models for wild and mutant type proteins (Figure 2). The de-stability of structural complexes due to free energy change of the built models indicated significant change in the structure of the protein that can hamper its natural function. Furthermore, active site analysis confirmed the structural variation and drug ineffectiveness against mutant model as active site amino acid Thr-493 does not interact with the drug (Figure 1).
In the purpose to identify diagnostic biomarker and prognostic significance, we analyzed the distribution of CC, CT and TT and the frequency of C and T allele among the study population. All the analysis done in this report was stratified according to gender. The genotypic distributions of three genotypes (TT, CT and CC) showed a good congruence with Hardy–Weinberg law, implying the consistent flow of these genotypes in Bangladeshi population including male and female groups. The χ2 values calculated for all genotypes of target SNP were satisfactory, even in patient and control subjects of the study population (Table S1).
The presence of CT genotypes increased the relative risk for T2DM by 1.91 fold among all the patients. In contrast, the ratio of risk to get T2DM rose to 3.03 × in the case of females who had the CT genotype. However, the male patients with the same genotype showed no association (Table 2). Interestingly, a study conducted in India had found the highest frequency for TT with significant p value where CT was less significant (Umapathy et al., 2012). Likewise, two studies conducted among the South and North Indian population found the association of diabetic foot ulcer with TT genotype and T allele (Umapathy et al., 2012; Zubair & Ahmad, 2018). However, another research on diabetic foot ulcer revealed no significant association with the polymorphism among Tamil Nadu populace of India (Mir et al., 2009). Our results are found to be consistent with an earlier report which revealed that T allele conferred a significant protection for Diabetic macroalbuminuria subjects (Dhamodharan et al., 2017).This variation between two adjacent populations indicated the transformation of ancestral T allele to C allele in the Bangladeshi population and this incorporation of C allele might have some effect on disease prognosis. The impact of the C allele over the T allele was also determined by odd ratio estimation and allele frequency analysis. The risk estimation analysis for allele variation yielded a similar result as assumed previously from genotypic-association analysis. The association of the C allele was observed in patients compared to the control group. The most significant association of the C allele was found for female patients indicating that the C allele might be a potential risk factor for T2DM development especially in female. But the OR and p value could not produce the considerable signal in case of male patients (Table 2). When we persued the cumulative effect of genotypes with stress and age on T2DM patients separately, it revealed that among the female patients, the effect of stress on T2DM incidence was genotype independent (Table 3) despite the fact that the CT genotype increased risk for T2DM independently (Table 2). On the other hand, existence of the CT genotype in male patients raised the probability of getting T2DM by about 4 × more than that of CC/TT genotypes presence (Table 3), though the CT genotype did not have an effect on male T2DM patients individually (Table 2). In the same way, the effect of age on T2DM prevalence was almost independent to genotype (Table 3), despite a gradual rise in T2DM incidence with the increasing age of patients (Table S4).
From this analysis, a strong association of C allele or the CT genotype with T2DM is evident. However, when we compared CC with TT, we found the values of the odds ratio (OR) and relative risk (RR) were maximum though the CC genotype was not significantly present in the Bangladeshi people. At the same time, we found that the OR and RR values for the CC/CT ratio were close to 1, suggesting that only one C allele in genotype is enough for exerting its effect on developing type 2 diabetes mellitus (Table S3).
In conclusion, our study shows that the CT genotype of HSP70-hom +2,437 C/T polymorphism represents a stress-independent risk factor for T2DM incidence among women. This study also suggests that under stressful condition, males are susceptible to T2DM in the presence of the CT genotype (Figure 3). To our knowledge, it is the first study that showed the cumulative effect of stress and rs2227956 polymorphism on T2DM onset. To generalize the association as the potential biomarker, the findings should be replicated through more studies with a large number of samples and in different populations.
ACKNOWLEDGMENT
The Authors are highly grateful to the ministry of science and technology (MOST), Government of the People's Republic of Bangladesh for their extended support to National Institute of Biotechnology (NIB) during this work.
CONFLICT OF INTEREST
The authors declare that they have no competing interest.
AUTHOR CONTRIBUTION
SH contributed to conception, study design, and sample collection of the project. MM contributed to the sample collection, data producing, data analysis, interpreting the findings, manuscript writing, reviewing, and editing of the study. IA contributed to the sample collection, data producing and analysis and draft preparation of manuscript. MUH performed bioinformatics analysis, collected sample and produced data. MSAM and SB collected sample and produced data. MHR, PKS, and KCD contributed to sample collection and reviewing the manuscript. MS contributed to conception and critically reviewed the manuscript.




