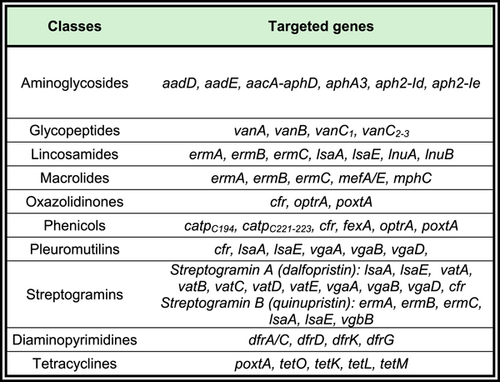
An in-house 45-plex array for the detection of antimicrobial resistance genes in Gram-positive bacteria
Graphical Abstract
Here we describe an in-house bead array targeting antimicrobial resistance (AMR) genes of Gram-positive bacteria and allowing their rapid detection all at once at reduced costs. A total of 27/41 AMR probes targeting genes frequently associated with resistance tested positive on a collection of 124 enterococci and 62 staphylococci isolated from healthy livestock animals. The array detected AMR genes associated with phenotypic resistance for 93% and 89.2% of the individual resistant phenotypes in enterococci and staphylococci, respectively.
Abstract
Identifying antimicrobial resistance (AMR) genes and determining their occurrence in Gram-positive bacteria provide useful data to understand how resistance can be acquired and maintained in these bacteria. We describe an in-house bead array targeting AMR genes of Gram-positive bacteria and allowing their rapid detection all at once at a reduced cost. A total of 41 AMR probes were designed to target genes frequently associated with resistance to tetracycline, macrolides, lincosamides, streptogramins, pleuromutilins, phenicols, glycopeptides, aminoglycosides, diaminopyrimidines, oxazolidinones and particularly shared among Enterococcus and Staphylococcus spp. A collection of 124 enterococci and 62 staphylococci isolated from healthy livestock animals through the official Belgian AMR monitoring (2018–2020) was studied with this array from which a subsample was further investigated by whole-genome sequencing. The array detected AMR genes associated with phenotypic resistance for 93.0% and 89.2% of the individual resistant phenotypes in enterococci and staphylococci, respectively. Although linezolid is not used in veterinary medicine, linezolid-resistant isolates were detected. These were characterized by the presence of optrA and poxtA, providing cross-resistance to other antibiotics. Rarer, vancomycin resistance was conferred by the vanA or by the vanL cluster. Numerous resistance genes circulating among Enterococcus and Staphylococcus spp. were detected by this array allowing rapid screening of a large strain collection at an affordable cost. Our data stress the importance of interpreting AMR with caution and the complementarity of both phenotyping and genotyping methods. This array is now available to assess other One-Health AMR reservoirs.
1 INTRODUCTION
Antimicrobial resistance (AMR) has become a major concern threatening public health (Nowakiewicz et al., 2019). For many decades, antibiotics are widely used in animal and human areas, leading to the worldwide resistance phenomenon. Indeed, bacteria have always been able to adapt by developing or acquiring mechanisms of resistance (Duval et al., 2019). In response to selective pressure and to survive antibiotic exposure, resistance occurs generally through the acquisition of genes located on mobile elements (Argudin et al., 2017; Strauss et al., 2015). Despite awareness associated with restrictive measures in animal production, the effects of the intensive use of drugs seem to persist lengthily, impacting numerous environments and ecological niches (Argudin et al., 2017; Nowakiewicz et al., 2019). Zoonotic, as well as commensal bacteria, became resistant providing them with a selective advantage (Argudin et al., 2017; Perreten et al., 2005) and sharing a potential risk of dissemination of AMR genes (De Jong et al., 2019; Nowakiewicz et al., 2019; Perreten et al., 2005).
Commensal bacteria, such as Enterococcus spp. are natural inhabitants of the gastrointestinal tract of healthy animals and humans. Enterococci can also be considered opportunistic pathogens when found related to human infections. They can persist in the environment and survive in different ecological niches such as soil, water, food items, and sewage (Nowakiewicz, 2019; Osman et al., 2019; Raza et al., 2018; Torres et al., 2018). Enterococci have emerged as one of the four most prevalent nosocomial human pathogens worldwide, especially due to their virulence, ability to form a biofilm, and intrinsic resistance (Leite-Martins et al., 2015; Raza et al., 2018). Notably, Enterococcus spp. are intrinsically resistant to a number of antimicrobials including trimethoprim-sulfamethoxazole, vancomycin (Enterococcus gallinarum, Enterococcus casseliflavus, and Enterococcus flavescens), streptogramins (Enterococcus faecalis) and exhibit low-level resistance to ß-lactams and aminoglycosides (Argudin et al., 2017; Torres et al., 2018; Zaheer et al., 2020). Since the 1980s, AMR enterococci have been the leading cause of hospital-acquired bloodstream and urinary tract infections, mainly through biofilm formation on catheters and implanted medical devices (Argudin et al., 2017; De Jong et al., 2019; Mercuro et al., 2018; Raza et al., 2018; Torres et al., 2018; Zaheer et al., 2020). Particularly, the majority of enterococcal infections are caused by E. faecalis and Enterococcus faecium, the most common species encountered in the human gut (Argudin et al., 2017; Mercuro et al., 2018; Raza et al., 2018; Torres et al., 2018). E. faecium has become a prominent cause of nosocomial infections often characterized by high-level resistance to multiple antibiotics (De Jong et al., 2019). Due to the high plasticity of enterococcal genomes, transfer and acquisition of AMR determinants in enterococci and other Gram-positive bacteria are then facilitated (Leite-Martins et al., 2015; Nowakiewicz et al., 2019).
Staphylococcus spp. are commensal bacteria widely found on the skin or mucosal surfaces of animals and humans (Alharbi, 2019; Bortolaia et al., 2016; Craft et al., 2019; Dastgheyb & Otto, 2015; Wendlandt et al., 2013). Despite preventing colonization by pathogenic bacteria (Alharbi, 2019), staphylococci are opportunistic pathogens often responsible for chronic and severe nosocomial infections (Bortolaia et al., 2016; Dastgheyb & Otto, 2015). Specifically, Staphylococcus aureus is one of the most pathogenic bacteria associated with human and animal diseases causing persisting skin and soft tissue infections (SSTIs), infectious endocarditis, septic arthritis, and osteomyelitis (Alharbi, 2019; Craft et al., 2019; Dastgheyb & Otto, 2015). The colonization of skin or mucosal surfaces by methicillin-resistant S. aureus (MRSA) and its dissemination in healthcare settings represents a global health issue (Holmes et al., 2015; Watkins et al., 2019) especially due to its capacity to acquire new AMR and to spread rapidly (Alharbi, 2019; Holmes et al., 2015). Indeed, methicillin resistance was first observed among clinical isolates before its rapid spread to the community (Turner et al., 2019). Staphylococcus spp. may exchange resistance determinants to numerous other bacteria in the same animal or human host, or between hosts by direct contact or through excretions such as sneezing, coughing, or licking (Wendlandt et al., 2013). Even if MRSA-associated infection rates have declined, these human infections are still problematic since they are characterized by broadening AMR and high rates of hospitalization and mortality (Purrello et al., 2016).
Over time, AMR has widely spread, consequently leaving healthcare personnel with last-line antibiotics (i.e., vancomycin, linezolid, and daptomycin) as the preferred means, and sometimes the only options to treat multidrug- resistant (MDR) infections (Raza et al., 2018; Sadowy, 2018; Torres et al., 2018; Zaheer et al., 2020). Resistance to last-resort drugs was already observed as well (Azhar et al., 2017; Doern et al., 2016; Holmes et al., 2015; Purrello et al., 2016) and among various bacteria, mainly reported in clinical settings (Argudin et al., 2017; Zaheer et al., 2020). Specifically, due to the alarming worldwide emergence (Osman et al., 2019; Raza et al., 2018), vancomycin-resistant enterococci (VRE) was ranked as a pathogen of high priority by the World Health Organization (WHO) (Wist et al., 2020). Due to its ability to confer high levels of vancomycin-resistance, vanA is the most reported gene among clinical VRE (Azhar et al., 2017; Turner et al., 2019; Watkins et al., 2019) and widely spread to other co-infecting bacteria such as S. aureus. Despite restricted use, resistance to linezolid has been reported in various species, strains, and settings (Sadowy, 2018). Determinants associated with linezolid resistance such as the highly mobilizable cfr coding for a ribosomal methyltransferase conferring cross-resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A or the oxazolidinone-phenicol transferable resistance gene optrA have been identified in staphylococci, enterococci and other Gram-positive species isolated from both humans and animals (Azhar et al., 2017; Doern et al., 2016; Sadowy, 2018; Timmermans et al., 2022a; Torres et al., 2018). The most recently described poxtA mediating decreased susceptibility to oxazolidinones, phenicols, and tetracyclines has also been detected in different Gram-positive bacteria of animal and human origin (Brenciani et al., 2018; Ruiz-Ripa et al., 2020; Sadowy, 2018; Timmermans et al., 2022a).
In summary, AMR is not restricted to a particular species or host. It became essential to consider all bacteria as a potential pool of resistance determinants possibly transferable to other pathogenic or commensal bacteria both in animals and humans (Argudin et al., 2017; De Jong et al., 2019; Osman et al., 2019; Perreten et al., 2005; Torres et al., 2018; Zaheer et al., 2020). Therefore, it is important to identify AMR determinants spreading in humans and animals (Perreten et al., 2005; Strauss et al., 2015) and to consider them as a One Health AMR pool. The complexity of AMR and particularly the cross-resistance phenomenon, that is resistance to multiple distinct antimicrobial classes conferred by a single molecular mechanism, requires monitoring all putative main sources of AMR at the genetic level.
We describe an in-house developed array targeting major AMR genes of Gram-positive bacteria and allowing their rapid and efficient detection all at once at reduced costs. In this study, we aimed to target AMR genes commonly found and particularly shared among Enterococcus and Staphylococcus spp. The diversity of probes per antimicrobial class reflects phenotypes observed during several years of field monitoring and pointed out the most critical (e.g., glycopeptides) or emergent ones (e.g., linezolid) to investigate. The presence of these AMR genes was studied in a collection of 124 enterococci and 62 staphylococci isolated from healthy livestock animals through the official Belgian AMR monitoring during the period 2018–2020, to decipher the genetic nature of this pool of AMR and thereby provide useful data to understand how resistance can be acquired and maintained in these bacteria.
2 MATERIALS AND METHODS
2.1 Isolate collection
All Enterococcus spp. and MRSA isolates included in this study were collected during the voluntary Belgian AMR monitoring programs of 2019 and 2020 and 2018 and 2019, respectively, from healthy food-producing animals by the Federal Agency for the Safety of the Food Chain (FASFC, 2020, http://www.favv.be/productionanimale/antibioresistance/resultats/#sciensano). Enterococcus spp. were isolated from fecal samples of pigs (N = 15 E. faecalis and 8 E. faecium), veal calves (N = 22 E. faecalis and 16 E. faecium), turkeys (N = 4 E. faecalis and 7 E. faecium) and broilers (N = 9 E. faecalis and 30 E. faecium) collected at a slaughterhouse and of chicken breeding hens (N = 4 E. faecalis and 6 E. faecium) and laying hens (N = 3 E. faecalis) collected at farm. MRSA have been isolated from nasal swab samples of sows (N = 26), fattening pigs (N = 25), or calves (N = 11) collected at the farm.
The selection of isolates was based on phenotypic resistance profiles determined by broth microdilution (BMD) assay carried out during the annual monitoring by Sciensano and interpreted according to European Food Safety Authority (EFSA) guidelines related to the European decision (2013/652/EU). Among selected staphylococci and enterococci isolates, all were multi-resistant and/or displayed phenotypic resistance to critical antibiotics such as vancomycin or linezolid. All selected MRSA were resistant to at least six different antimicrobial classes, including β-lactams (i.e., penicillin and cefoxitin). Resistance to antimicrobials included in both Enterococcus spp. and S. aureus testing panels (EUVENC and EUST plates, SensititreTM; Thermo Fisher Scientific) was targeted by this array (see Table 1) and directed isolate selection. Most targeted resistance genes are shared by both enterococci and staphylococci. Control isolates (N = 25) originated from the European Reference Laboratory for Antimicrobial Resistance (EURL-AR), from Sciensano internal collection, or other sources as listed in Table A1.
| Classes | Antimicrobials | Targeted genes |
|---|---|---|
| Aminoglycosides | Kanamycina1 (KAN) | aadDa, aadEc, aacA-aphDab, aphA3a, aph2-Idab, aph2-Iebc |
| Gentamicinb (GEN) | ||
| Streptomycinc1 (STR) | ||
| Glycopeptides | Vancomycin (VAN) | vanA, vanB, vanC1, vanC2-3 |
| Lincosamides | Clindamycin1 (CLN) | ermA, ermB, ermC, lsaA, lsaE, lnuA, lnuB |
| Macrolides | Erythromycin (ERY) | ermA, ermB, ermC, mefA/E, mphC |
| Oxazolidinones | Linezolid (LZD) | cfr, optrA, poxtA |
| Phenicols | Chloramphenicol (CHL) | catpC194, catpC221-223, cfr, fexA, optrA, poxtA |
| Pleuromutilins | Tiamulin1 (TIA) | cfr, lsaA, lsaE, vgaA, vgaB, vgaD, |
| Streptogramins | Quinupristin (Group B—streptogramin)/dalfopristin (Group A—streptogramin) (Synercid [SYN]) | Streptogramin A (dalfopristin): lsaA, lsaE, vatA, vatB, vatC, vatD, vatE, vgaA, vgaB, vgaD, cfr Streptogramin B (quinupristin): ermA, ermB, ermC, lsaA, lsaE, vgbB |
| Diaminopyrimidines | Trimethoprim1 (TMP) | dfrA/C, dfrD, dfrK, dfrG |
| Tetracyclines | Tetracycline (TET) | poxtA, tetO, tetK, tetL, tetM |
- 1Antimicrobials monitored by broth microdilution in Staphylococcus spp. only.
2.2 Antimicrobial susceptibility testing (AST)
Antimicrobial minimum inhibitory concentrations (MICs) were determined by BMD with EUVENC (for enterococci) and EUST (for staphylococci) multiwell plates (Sensititre™; Thermo Fisher Scientific) and interpreted according to the EFSA guidelines as detailed in ad hoc reports (Federal Agency for the Safety of the Food Chain, 2020, http://www.favv.be/productionanimale/antibioresistance/resultats/#sciensano) and based on epidemiological cut-off values (ECOFFs). Antimicrobial susceptibility testing (AST) for clindamycin, kanamycin, streptomycin, tiamulin, and trimethoprim was performed in S. aureus only.
2.3 AMR genes detection with the in-house array
The molecular method developed for the detection of AMR genes is a multiplex assay based on a Ligase Chain Reaction (LCR) of Padlock-shaped Probes (PLPs) followed by hybridization of LCR products with MagPlex-TAG™ microspheres coated with unique 24-nt long capture tags and detection with a Luminex® 200™ instrument (Luminexxas).
2.3.1 PLPs probes design
Probes were designed as described previously (Boland et al., 2018; Timmermans et al., 2022b; Wattiau et al., 2011). The sequences of the probes developed in this study and used in the LCR assay and the corresponding AMR genes targeted by the array are listed in Table 2. Their design was based on published DNA sequences of AMR genes from the NCBI nucleotide database (https://www.ncbi.nlm.nih.gov/nuccore). Several sequences of the same AMR gene from both Enterococcus and Staphylococcus spp. were aligned with Bionumerics 6.6 (bioMérieux SA) and target-specific sequences were selected within the most conserved regions. In addition to these specific sequences corresponding to the two extremities of the probes, the so-called 5'-arms and 3’-arms, probes encompassed: sequences of the universal primers “reverse” (cUR) and “forward” (UF; see Table 2) as well as an anti-TAG sequence matching a given TAG sequence of the MagPlex-TAG™ microspheres (Luminex). Probes are schematically represented in linear form as follows: 5'arm – cUR – AA – UF – anti-TAG – 3'arm where “AA” is a di-deoxyadenosine linker.
| Probes | MagPlex-TAG™ microspheres | Final conc. pMb | Sequence |
|---|---|---|---|
| aacA-aphD | MTAG-A022 | 400 | TTTGCCAGAACATGAATTACACGAGGG-cUR-AA-UF-CAAACAAACATTCAAATATCAATCACCAAAAATCTGGTTTTAGAATTATTGAAGA |
| aadD | MTAG-A047 | 200 | GTCTGACGACCAAGAGAGCCATAAACA-cUR-AA-UF-TCTCTTTAAACACATTCAACAATAATATCCGAATAGGGCCCATCA |
| aadE | MTAG-A056 | 200 | TCATGTAAGAAAGCCAAGCGCAAGG-cUR-AA-UF-CTTAAACTCTACTTACTTCTAATTGGGACATAGTTCCGACTGATATAGATTA |
| aph2-Id/aph2-Ie | MTAG-A027 | 200 | TGCAGCTATTTCTGATCCCGACAATG-cUR-AA-UF-TAACTTACACTTAACTATCATCTTCATTTGTGGAATAATCGATTTTGGAGA |
| aphA3 | MTAG-A028 | 200 | TGGCTGGAAGGAAAGCTGCC-cUR-AA-UF-CACTTAATTCATTCTAAATCTATCACGGGAAAAGGACATGATGCTA |
| catpC194 | MTAG-A021 | 400 | ACTGGTTACAATAGCGACGGAGAG-cUR-AA-UF-TCAAACTCTCAATTCTTACTTAATGGTGATAAACTCAAATACAGCTTTTAGA |
| catpC221,/catpC223 | MTAG-A026 | 200 | CCATACCGATTTCAATGATTCCTTGGATTG-cUR-AA-UF-TACATTCAACACTCTTAAATCAAACCTAAAAAACCGATACCTGAAAACA |
| cfr | MTAG-A020 | 200 | TGCAAACGAAGTTGTTAGCCTTCTTAAAAG-cUR-AA-UF-CTTTCTCATACTTTCAACTAATTTGGTGTAAATGATTCTCTTGAGCA |
| dfrA/dfrC | MTAG-A034 | 200 | ACCAAGCTTCATTTCACCATGAAGGG-cUR-AA-UF-ACTTATTTCTTCACTACTATATCAAGACGTAACGTCGTACTCACTA |
| dfrD | MTAG-A035 | 200 | ATCGGAAGGGCTTTACCTGACAGAA-cUR-AA-UF-CATCTTCATATCAATTCTCTTATTAATCATATTAGGTAGAAAGAACCTTCAATCA |
| dfrG | MTAG-A044 | 400 | GAATGACATTCCTTGGAGGATTCCCAA-cUR-AA-UF-TCATCACTTTCTTTACTTTACATTGATAAGAATAGAGTGATTGGCAAAGA |
| dfrK | MTAG-A042 | 200 | TCTTGAATCAATCGGAAGAGCA-cUR-AA-UF-CACTACACATTTATCATAACAAATAGGGATATCCAATTATATTAGGAAGGAAGAA |
| ermA | MTAG-A046 | 800 | TTTGCATGCTTCAAAGCCTGTCG-cUR-AA-UF-TTAAACAATCTACTATTCAATCACTCCTTCGATAGTTTATTAATATTAGTGACA |
| ermB | MTAG-A061 | 200 | AGTCTCGATTCAGCAATTGCTTAAGCT-cUR-AA-UF-AATCTCTACAATTTCTCTCTAATAGTTGCTCTTGCACACTCA |
| ermCa | MTAG-A014 | 200 | TTAGTACAGAGGTGTAATTTCGTAACTGCC-cUR-AA-UF-AATTTCTTCTCTTTCTTTCACAATGAAAAGGSCATTTTACCCTTGAA |
| fexA | MTAG-A029 | 200 | TGATATTGTTGCCAGGTGGTGTGG-cUR-AA-UF-TACTACTTCTATAACTCACTTAAACTTCTGGACAGGCTGGAA |
| Gram+ | MTAG-A052 | 200 | CCTTCCTCCGGTTTGTCACC-cUR-AA-UF-TTCTTCATTAACTTCTAATCTTACATGATTTGACGTCATCCCCA |
| lnuA | MTAG-A074 | 800 | GATATAGATTTTGACGCTCAACACACTCAA-cUR-AA-UF-ACACTCATTTAACACTATTTCATTAAACAACAAAGAGAACACAGAGATATA |
| lnuB | MTAG-A075 | 400 | TGGATCGTTTACCAAAGGAGAAGGTGAC-cUR-AA-UF-CATAAATCTTCTCATTCTAACAAAATGAACGAATTACAGCTTGTATGATGTA |
| lsaA | MTAG-A036 | 400 | TTGACGGTGGAAGAGCTTCGTC-cUR-AA-UF-ATTAAACAACTCTTAACTACACAAGCCAACGCATCACAAAACATTA |
| lsaE | MTAG-A012 | 400 | TGTTGGACATAAGGCTGCAAAAGCG-cUR-AA-UF-CATAATCAATTTCAACTTTCTACTCTGGTTCAAAACTGGATAAGGGTTA |
| mefA/mefEa | MTAG-A045 | 200 | GCAATTGTACGTATWCCTAAGCTGGGT-cUR-AA-UF-TACACAATATTCATCATAACTAACGCTGTGATTGCATCTATTACGGTA |
| mphC | MTAG-A063 | 200 | TCGCTGAGTTTGCTATGGAATCAGGAG-cUR-AA-UF-CTAAATCACATACTTAACAACAAAACTCAATGCAGTATTCCCAATGTTTA |
| optrA | MTAG-A018 | 400 | GAAGGAGAAGGTTAAGAAGGAGAAACG-cUR-AA-UF-ACACTTATCTTTCAATTCAATTACGCGATCGTAACTCCATTGA |
| poxtA | MTAG-A055 | 200 | CCAGTGGAAATGCCCGTATTGGTTAT-cUR-AA-UF-ACATCAAATTCTTTCAATATCTTCCTTGAACTTGATAATGGTTCACTGA |
| sodA-fm C | MTAG-A054 | 800 | AGACAGCTGTACGTAACAATGGTGG-cUR-AA-UF-TTAATACAATTCTCTCTTTCTCTAGGACGCTATTCCAACAGATATCA |
| sodA-fs C | MTAG-A057 | 800 | TGGTGGCGGTCACGC-cUR-AA-UF-ACTTACAATAACTACTAATACTCTCGTACAGCCGTTCGTAACAA |
| staph C | MTAG-A053 | 200 | TGCTACGGTGAATACGTTCCCG-cUR-AA-UF-TTAACAACTTATACAAACACAAACTCGCTAGTAATCGTAGATCAGCA |
| tetK | MTAG-A066 | 200 | TTCCCTTCACTGATTATGGTGGTTGTAGC-cUR-AA-UF-TCTTACTAATTTCAATACTCTTACAGGAGTAGGATCTGCTGCA |
| tetL | MTAG-A067 | 400 | GCTAAGTACTGCCGAAATCGGAAGTG-cUR-AA-UF-ATCTCAATTACAATAACACACAAAGTTCCTTATATGATGAAAGATGTTCACCA |
| tetM | MTAG-A072 | 400 | AAGGTACAACGAGGACGGATAATACGC-cUR-AA-UF-CTATCATTTATCTCTTTCTCAATTCAGAATTAGGAAGCGTGGACA |
| tetO | MTAG-A065 | 200 | GACAGCAGTGACATCTTTTCAGTGGG-cUR-AA-UF-TACTTAAACATACAAACTTACTCAGTCAAAGGGGAATCACTATCCA |
| vanA | MTAG-A013 | 200 | AGTCAATACTCTGCCCGGT-cUR-AA-UF-CAAATACATAATCTTACATTCACTGCCGCATTGTACTGAACGA |
| vanB | MTAG-A033 | 200 | ATGCGGGCATCGCCG-cUR-AA-UF-ACTACTTATTCTCAAACTCTAATATCACTGGCCTACATTCTTACAAAAA |
| vanC1 | MTAG-A030 | 200 | TGGGAATCGCTAGTGCTCCCAC-cUR-AA-UF-CTTAACATTTAACTTCTATAACACTCTTGCATCAACTTGCTGATACCA |
| vanC2/vanC3a | MTAG-A051 | 200 | TCGAAGCACTCCAATCATCTCCC-cUR-AA-UF-CAATTTACATTTCACTTTCTTATCTTAGCYTCAGCAACTAGCGCAA |
| vatA | MTAG-A073 | 200 | TTGCTGCAGAAGCTGTTGTCACAAAG-cUR-AA-UF-CTTTATCAAATTCTAATTCTCAACGGGACGGGGCAATCA |
| vatB | MTAG-A039 | 200 | ATTCAAATAGGAGATGGAGCAATTGTTGCT-cUR-AA-UF-ACAAATATCTAACTACTATCACAAAGAATGTTACTGTTATGCCAGGA |
| vatC | MTAG-A078 | 200 | ATGGTTGGGAGAAGCATACCCCTA-cUR-AA-UF-TTTACAAATCTAATCACACTATACCAACATTTCCATTCAATCTTTTCGGAA |
| vatD | MTAG-A077 | 200 | GCGCCATACATGTTAGCTGGAGGAA-cUR-AA-UF-AATAACAACTCACTATATCATAACGCTGCTAATTCTGTTGTTGTAAAAGATATA |
| vatE | MTAG-A076 | 200 | CCGATTTTGAGAAACACGTTACCCATCAC-cUR-AA-UF-TCTCATCTATCATACTAATTCTTTCTATTATGATGACCCAGTAAATCCCA |
| vgaA | MTAG-A025 | 200 | CTCAGTCGATTGAGTATTGAACCTTCGGAA-cUR-AA-UF-CTTTCTTAATACATTACAACATACACTTTTACTTGAGACAAAAATTACAGAAGTA |
| vgaB | MTAG-A038 | 200 | ATAAGGCGCAAGGAATGATTAAGCCC-cUR-AA-UF-ATTCAATACTATCTAACACTTACTGGAGCAAGCTATAAAGCTAAAAGAGA |
| vgaDc | MTAG-A043 | 200 | TTGTTTGCCGACGAACCAACTACAAAC-cUR-AA-UF-AACTTTCTCTCTCTATTCTTATTTCATTAGCGATGAAGGCGGAAATA |
| vgbB | MTAG-A048 | 200 | TCACTAGTGGTAACGATGGTGCAC-cUR-AA-UF-AATCAACACACAATAACATTCATAAGCGGCTCCAGTGGGTA |
| UF | GTAGACTGCGTACCAATTC | ||
| UR | GACGATGAGTCCTGAGTAA | ||
| cUR | TTACTCAGGACTCATCGTC |
- Note: Nucleotide sequence of the probe (from 5' to 3'). Bold characters highlight the sequence targeted on the template DNA, underlined characters indicate the sequence complementary to the Luminex® MagPlex-TAG™microspheres with the bead ID indicated in the second column; UF, UR, and cUR, nucleotide sequence of the PCR amplification primers Universal Forward, Reverse and complementary UR, respectively (Wattiau et al., 2011; Boland et al., 2018); normal characters indicate nucleotides added to reach a final set of probes with evenly distributed sizes ranging from 99 to 125 nucleotides. For the design of each probe, the most conserved region of the targeted gene, obtained from alignments in Bionumerics© of sequences available in GenBank, was chosen as the target sequence. Three control probes (indicated with the letter “C”) were designed for the identification of each bacterial species, Enterococcus faecalis, Enterococcus faecium, and Staphylococcus aureus.
- a Nucleotide sequence including a wobble.
- b Final concentration in the first step of the Ligase Chain Reaction (LCR) assay.
- c No control was available to validate this PLP.
2.3.2 DNA preparation
The DNA samples were prepared from bacteria grown on Columbia Sheep Blood agar plates (Oxoid) with the QIAGEN DNeasy blood and tissue kit (Qiagen) according to the manufacturer's instructions for Gram-positive bacteria. Both DNA purity and concentration were assessed using the Nanodrop 1000 (Isogen Life Science) before storage at −20°C.
2.3.3 LCR assay
The LCR assay was conducted in three successive steps according to the procedure of Boland et al. (2018) with minor modifications. The first step (ligation) was conducted in a 10 µL mixture containing 1 µL of Pfu DNA ligase buffer 10X (#600191-52; Agilent), 2 U of Pfu DNA ligase (#600191-51; Agilent), a specific final concentration of each PLP detailed in Table 2 and 1 µL of DNA (≥10 ng μL−1) extracted as described above. The thermoCycler conditions were as follows: denaturation at 95°C for 3 min followed by 25 cycles of 30 s at 95°C and 5 min at 65°C and 2 min at 98°C for final denaturation. The second step (exonuclease treatment) consisted of the addition of 15 µl of exonuclease mixture containing 67 mM glycine- KOH, pH 9.4, 2.5 mM MgCl2, 50 μg mL−1 bovine serum albumin (BSA) (#B0262S; New England BioLabs) and 0.0015 U λ exonuclease (#M262S; New England BioLabs) to the step 1 LCR products. The resulting 25-µL sample was incubated at 37°C for 45 min followed by a 10 min inactivation at 95°C. As the third step, PCR amplification was performed by adding a mix of 50 µL of 2x Absolute qPCR mixture (Thermo Scientific) supplemented with 2 µL of universal reverse UR primer concentrated at 2.5 µM and 2 µL 5'Cy3-labeled universal forward UF primer concentrated at 20 µM to each sample. After 10 min denaturation at 95°C, 30 cycles of 45 s at 95°C, 45 s at 55°C and 1 min at 72°C were conducted and followed by 15 min of final elongation at 72°C and 2 min of denaturation at 98°C. LCR products were stored at −20°C until hybridization and reading on a bead array platform.
2.3.4 Hybridization and detection of the LCR products on a bead-array platform
A bead mix was prepared in TE buffer pH 8 to obtain a final concentration of 1 x105 beads per mL per region of each MagPlex-TAG™ microsphere listed in Table 2 and an additional bead unrelated to the probes and used as negative hybridization control. The bead mix was stored at 4°C and protected from light.
Before hybridization, the bead mix was pelleted on a magnet and suspended in 2× hybridization buffer (0.4 M NaCl, 0.2 M Tris, 0.16% Triton X-100, pH 8.0) at a concentration of 50 beads of each type per µL. The hybridization step consisted in mixing 25 µL of the bead mix and 25 µL of the final LCR product followed by 90 s of denaturation at 96°C and 30 min of hybridization at 37°C.
Right after hybridization, three washes were performed by pelleting the beads on a magnetic bead separation system (V&P Scientific) for 1 min, removing the supernatant by forceful inversion, and suspending the beads in 75 µl 1× hybridization buffer (0.2 M NaCl, 0.1 M Tris, 0.08 Triton X-100, pH 8.0). After the final wash, the plate was incubated at 37°C for 15 min in the Luminex® 200™ instrument.
Bead-array analysis was performed on 50 µL of the final solution at 37°C and Median Fluorescence Intensity (MFI) was measured on at least 100 beads of each bead type.
2.3.5 Data analysis
The fluorescence signal of the Gram-positive control probe “Gram+” was used as an internal standard to normalize MFI signals observed for each sample according to the formula: (MFI probe/MFI Gram+) × 100. The results expressed in normalized MFI (nMFI) were evaluated against cut-offs. Cut-offs were determined after plotting experimental nMFI values obtained by testing isolates and control strains and included a twofold ratio between positive and negative nMFI. The probes of the array were validated with reference strains used as positive controls, except for the vgaD probe for which no reference strain was available (see Table A1). The control probes soda_fs and soda_fm are based on the housekeeping sodA gene of E. faecalis and E. faecium, respectively; the staph control probe is based on the 16S rRNA gene of S. aureus. Control probes were validated on E. faecalis ATCC 29212, E. faecium 2013/16227 field strain, and S. aureus ATCC 29213 reference strains. These three control strains together with the negative control Escherichia coli ATCC 25922 were used in each experiment to validate the LCR assay.
2.4 Next-generation sequencing analysis
A total of 31 isolates (16 E. faecalis, 14 E. faecium, and 1 MRSA) were analyzed by whole-genome sequencing to further investigate linezolid and vancomycin-resistant isolates and a few isolates resistant to five or more antimicrobials. Isolate selection was based on the resistance phenotypes, irrespective of the bacterial species, that is all isolates resistant to linezolid (11 E. faecalis and 11 E. faecium) and vancomycin (n = 2 E. faecalis). The remaining isolates (3 E. faecalis, 3 E. faecium, and 1 MRSA) were selected because of their MDR phenotype (resistant to at least five different antimicrobials). Genomic DNA was extracted as detailed in “DNA preparation.” Libraries were prepared using Nextera XT DNA library preparation kit (Illumina) according to the manufacturer's instructions and underwent Illumina sequencing using the MiSeq V3 chemistry (Illumina) for the production of two ×250 bp paired-end reads. Raw sequenced reads were trimmed with Trimmomatic v0.38 using default settings and de novo assembled with SPAdes v 3.13.0 using default settings (Bankevich et al., 2012). Identification of AMR genes was assessed with ResFinder 4.1 using default settings (Bortolaia et al., 2020).
3 RESULTS
In this study, a bead array based on AMR genes was developed and validated on reference strains (see Table A1) to characterize the genetic nature of resistance expressed by Gram-positive bacteria. This array was used to detect AMR genes in a selection of enterococci and staphylococci isolated from livestock animals in Belgium. A total of 186 Gram-positive bacteria including 57 E. faecalis, 67 E. faecium, and 62 MRSA were screened for the presence of the AMR genes detailed in Table 1. Altogether, 27 out of the 41 AMR probes tested positive on the 186 selected animal isolates (see Tables 3 and 4). In most cases, resistance profiles correlated with markers detected with the array. Results of this AMR gene screening are presented and discussed hereafter per antimicrobial class, followed by whole genome sequencing (WGS) analysis results conducted to further investigate linezolid and vancomycin-resistant isolates and a few isolates resistant to five or more antimicrobials.
| AMR genes | R-E. faecalis | R-E. faecium | R-S. aureus | AMR genes | R-E. faecalis | R-E. faecium | R-S. aureus |
|---|---|---|---|---|---|---|---|
| Chloramphenicol (CHL) | 33 | 13 | 12 | Quinupristin/dalfopristin (SYN) continued | 57 | 53 | 37 |
| catpC194 | 0 (0.0%) | 2 (15.4%) | 0 (0.0%) | lsaE | 20 (35.1%) | 37 (69.8%) | 29 (78.4%) |
| catpC221/223 | 4 (12.1%) | 1 (7.7%) | 0 (0.0%) | vatA | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| cfr | 0 (0.0%) | 0 (0.0%) | 1 (8.3%) | vatB | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| fexA | 17 (51.5%) | 5 (38.5%) | 9 (75.0%) | vatC | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| optrA | 18 (54.4%) | 8 (61.5%) | 0 (0.0%) | vatD | 1 (1.8%) | 0 (0.0%) | 0 (0.0%) |
| poxtA | 1 (3.0%) | 7 (53.8%) | 0 (0.0%) | vatE | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Clindamycin (CLN) | NT | NT | 57 | vgaA | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| ermA | – | – | 3 (5.3%) | vgaB | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| ermB | – | – | 15 (26.3%) | vgaD | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| ermC | – | – | 13 (22.8%) | vgbB | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| lsaA | – | – | 0 (0.0%) | ||||
| lsaE | – | – | 29 (50.9%) | Streptomycin (STR) | NT | NT | 11 |
| lnuA | – | – | 1 (1.8%) | aadE | – | – | 4 (36.4%) |
| lnuB | – | – | 26 (45.6%) | aph2-Id/Ie | – | – | 0 (0.0%) |
| Erythromycin (ERY) | 52 | 54 | 36 | Tetracycline (TET) | 53 | 65 | 61 |
| ermA | 12 (23.1%) | 25 (46.3%) | 3 (8.3%) | poxtA | 1 (1.9%) | 12 (18.5%) | 0 (0.0%) |
| ermB | 49 (94.2%) | 49 (90.7%) | 15 (41.7%) | tetK | 0 (0.0%) | 0 (0.0%) | 42 (68.9%) |
| ermC | 0 (0.0%) | 0 (0.0%) | 13 (36.1%) | tetL | 41 (77.4%) | 51 (78.5%) | 23 (37.7%) |
| mefA/E | 0 (0.0%) | 1 (1.9%) | 0 (0.0%) | tetM | 47 (88.7%) | 65 (100.0%) | 59 (96.7%) |
| mphC | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | tetO | 9 (17.0%) | 0 (0.0%) | 0 (0.0%) |
| Gentamicin (GEN) | 25 | 12 | 19 | Tiamulin (TIA) | NT | NT | 35 |
| aacA-aphD | 25 (100.0%) | 12 (100.0%) | 17 (89.5%) | cfr | 1 (2.9%) | ||
| aph2-Id/Ie | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | lsaA | – | – | 0 (0.0%) |
| Kanamycin (KAN) | NT | NT | 19 | lsaE | – | – | 29 (82.9%) |
| aacA-aphD | – | – | 17 (89.5%) | vgaA | – | – | 0 (0.0%) |
| aadD | – | – | 13 (68.4%) | vgaB | – | – | 0 (0.0%) |
| aph2-Id/Ie | – | – | 0 (0.0%) | vgaD | – | – | 0 (0.0%) |
| aphA3 | – | – | 0 (0.0%) | Trimethoprim (TMP) | NT | NT | 55 |
| Linezolid (LZD) | 11 | 11 | 0 | dfrA/C | – | – | 1 (1.8%) |
| cfr | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | dfrD | – | – | 0 (0.0%) |
| optrA | 11 (100.0%) | 9 (81.8%) | 0 (0.0%) | dfrG | – | – | 25 (45.5%) |
| poxtA | 0 (0.0%) | 8 (72.7%) | 0 (0.0%) | dfrK | – | – | 23 (41.8%) |
| Quinupristin/dalfopristin (SYN) | 57 | 53 | 37 | Vancomycin (VAN) | 2 | 0 | 0 |
| ermA | 12 (21.1%) | 20 (37.7%) | 3 (8.1%) | vanA | 1 (50.0%) | 0 (0.0%) | 0 (0.0%) |
| ermB | 49 (86.0%) | 35 (66.0%) | 6 (16.2%) | vanB | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| ermC | 0 (0.0%) | 0 (0.0%) | 7 (18.9%) | vanC1 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| lsaA | 57 (100.0%) | 0 (0.0%) | 0 (0.0%) | vanC2-3 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
- Note: For each antimicrobial, the numbers in the first line correspond to the number of isolates that were phenotypically resistant to this antimicrobial and assessed with the array. In front of each gene, the numbers in each column indicate the number of isolates in which this gene was detected with the array and the corresponding percentage of isolates carrying this gene among the number of isolates indicated in the top line for each antimicrobial. A dash indicates that the presence of the gene in the corresponding bacterial species was not assessed because the phenotypic resistance profile was not available.
- Abbreviation: NT, not tested.
| AMR genes | E. faecalis (n = 57) | E. faecium (n = 67) |
|---|---|---|
| Clindamycin (CLN) | ND | ND |
| lnuA | 0 | 0 |
| lnuB | 19 | 38 |
| Kanamycin (KAN) | ND | ND |
| aacA-aphD | 26 | 13 |
| aadD | 1 | 2 |
| aph2-Id/Ie | 0 | 0 |
| aphA3 | 35 | 24 |
| Streptomycin (STR) | ND | ND |
| aadE | 33 | 42 |
| aph2-Id/Ie | 0 | 0 |
| Trimethoprim (TMP) | ND | ND |
| dfrA/C | 0 | 0 |
| dfrD | 1 | 0 |
| dfrG | 35 | 26 |
| dfrK | 0 | 2 |
- Abbreviation: ND, not determined.
3.1 Streptogramins resistance
Quinupristin/dalfopristin (Q/D; a streptogramins B and A combination) resistance is correlated with the presence of lsa (for Lincosamides Streptogramins A resistance) genes family. Particularly, intrinsic in E. faecalis, this resistance is conferred by expression of the lsaA gene (Frye & Jackson, 2013; Hollenbeck & Rice, 2012; Singh et al., 2002; Torres et al., 2018), found in all E. faecalis (n = 57/57) of this study. Concordant with this specificity, lsaA was not detected in E. faecium nor S. aureus isolates. The second gene lsaE was found in 69.8% (n = 37/53) of Q/D-resistant E. faecium and 35.1% of E. faecalis (n = 20/57). Enterococcus faecalis isolates resistant to Q/D (n = 57) carried ermA (n = 2), ermB (n = 23), ermA/ermB (n = 5), ermB/vatD (n = 1), ermB/lsaE (n = 15) or ermA/ermB/lsaE (n = 5) in addition to lsaA, or lsaA only (n = 6). Enterococcus faecium isolates resistant to Q/D (n = 53) carried lsaE (n = 1), ermB (n = 7), lsaE/ermB (n = 18), lsaE/ermA (n = 1), ermA/ermB (n = 3), lsaE/ermA/ermB (n = 16) or lsaE/ermB/mefA/E (n = 1). vgaA, vgaB, vgaD, vatA, vatB, vatC, vatE, and vgbB, are also involved in streptogramins A or B resistance (see Table 1) (Cho et al., 2020; De Graef et al., 2007; Pechère, 2001; Petinaki & Papagiannitsis, 2019; Roberts, 2008) were screened with this array, but not detected. One of the vat genes, namely vatD, was identified in one E. faecalis isolate (VAR-683) resistant to erythromycin and Q/D. Note, vatD is reported in the literature to confer resistance to streptogramins A in Enterococcus spp. and being colocated with ermB (Hammerum et al., 2001; Jackson et al., 2007; Rende-Fournier 1993). In contrast with this and according to WGS analysis, vatD in VAR-683 was located on a distinct contig as compared to ermB. No resistance gene was detected in 6 Q/D-resistant enterococcal isolates. In staphylococci, 37 Q/D-resistant isolates were tested with the array and carried lsaE (n = 22), ermA (n = 2), ermC (n = 2) only or a combination of streptogramins A and/or B resistance genes: lsaE/ermB (n = 4), lsaE/ermC (n = 3), ermB/ermC (n = 1) and ermA/ermB/ermC (n = 1). lsaA, vga, vgb, and vat genes were tested but not detected in staphylococci. Two Q/D-resistant staphylococcal isolates harbored none of the investigated genes.
Noteworthy, the Q/D resistance mechanisms are still not perfectly understood. Indeed, few studies reported that resistance to streptogramin A is sufficient to confer resistance to Q/D (Hancock, 2005; Yan et al., 2021) while others support that a combination of both resistance to streptogramin A (vat or vga family) and streptogramin B (erm family or vgbB) is required to ensure Q/D resistance (Miller et al., 2014; Zarrouk 2000). Both lsaA and lsaE are the only genes reported to confer resistance to both Q/D components (Alcock et al., 2020; Singh et al., 2002; Wendlandt et al., 2013). In this study, the presence of lsaA, lsaE, or a combination of both streptogramin A and B resistance genes has been considered concordant with a Q/D resistance phenotype.
3.2 Macrolide resistance
Target modification by erm genes (coding for 23S rRNA methylases) is known to confer macrolide, lincosamide, and streptogramin B (MLSB) resistance and is the most common erythromycin resistance mechanism (Frye & Jackson, 2013; Jensen et al., 1999; Marosevic et al., 2017; Petinaki & Papagiannitsis, 2019; Roberts, 2008; Schwarz et al., 2018; Torres et al., 2018). In our study and as in Frye & Jackson (2013), ermA and ermB were both detected in enterococci, whereas all three ermA, ermB, and ermC were found in staphylococci. Among the three ermA, ermB, and ermC genes, ermB was the most observed in our study, with 92.5% of all erythromycin-resistant enterococcal isolates carrying this gene (n = 98/106) as reported in other studies (Frye & Jackson, 2013; Zaheer et al., 2020). Also, ermA was detected in 34.9% (n = 37/106) and found in combination with ermB in 30.2% of erythromycin-resistant isolates (n = 32/106). Furthermore, ermA was more frequently found in E. faecium (46.3% (n = 25/54) vs. 23.1% in E. faecalis (n = 12/52)) whereas ermB was identified equally among enterococcal isolates (94.2% E. faecalis [n = 49/52] and 90.7% E. faecium [n = 49/54]). Besides, neither ermC nor mphC was detected in the investigated enterococci isolates. In addition, mefA/E involved in macrolide resistance was targeted in this array and found in one E. faecium isolate resistant to erythromycin (MIC > 128 mg L−1) and Q/D (MIC = 2 mg L−1), characterized by the presence of ermB, lsaE, and mefA/E. mefA/E was first described in Streptococcus pneumoniae to confer macrolide resistance and was also reported in Enterococcus spp. from animals and humans (Petsaris et al., 2005). However, the presence of mefA/E has been reported to result in low erythromycin MICs (2–16 mg L−1) as compared to higher MICs (≥128 mg L−1) associated with erm genes (Pechère, 2001). In the particular case of a combination of ermB and mefA/E as observed in this study, inference of erm and mef genes based on MICs is not possible. No gene was detected in three erythromycin-resistant enterococcal isolates. In staphylococci, erythromycin resistance was reported in 36/62 isolates and characterized by ermA in 5.5% (n = 2), ermB in 36.1% (n = 13), ermC in 30.6% (n = 11), ermB/ermC in 2.8% (n = 1), and ermA/ermB/ermC in 2.8% (n = 1) of resistant isolates. mefA/E and mphC conferring macrolide resistance were not detected in staphylococci. In eight of these erythromycin-resistant isolates, no gene was detected.
3.3 Lincosamide resistance
While lincosamides (e.g., clindamycin) are not included in the susceptibility testing panel for enterococci, some genes targeted by the array confer resistance to this antimicrobial class: the lsa, vga, and erm genes mentioned above and the lnuA and lnuB genes (EFSA; Cho et al., 2020; Lozano et al., 2012). Intrinsic clindamycin resistance is conferred by the presence of lsaA in all E. faecalis (n = 57). Note, lnuB was detected in 33.3% and 56.7% of all E. faecalis (n = 19/57) and E. faecium (n = 38/67), respectively. In addition, lnuA, originally described in staphylococci (Cho et al., 2020; De Graef et al., 2007; Frye & Jackson, 2013) was not detected in enterococci. In staphylococci, several gene combinations were observed among clindamycin-resistant isolates (n = 57) namely, lsaE (n = 4), lnuA (n = 1), lnuB (n = 2), ermA (n = 2), ermB (n = 9), ermC (n = 8), ermB/ermC (n = 1), ermA/ermB/ermC (n = 1), lsaE/ermB (n = 1), lsaE/lnuB (n = 18), lsaE/ermB/lnuB (n = 3), and lsaE/ermC/lnuB (n = 3). One isolate harbored lnuA only but did not express clindamycin resistance (MIC = 0.25 mg L−1, just below the epidemiological cut-off value (ECOFF)). Four clindamycin-resistant isolates did not harbor any relevant gene.
3.4 Pleuromutilin resistance
Tiamulin resistance reported in 35 S. aureus is known to be conferred by lsa, cfr, or vga genes (Feßler et al., 2018; Schwarz et al., 2018; Van Duijkeren et al., 2014). In this study, 82.9% of tiamulin-resistant isolates harbored lsaE (n = 28) alone or in combination with cfr (n = 1), while lsaA and vga genes were not detected. Six tiamulin-resistant isolates did not harbor any relevant gene. AST for tiamulin resistance was not assessed in enterococci.
3.5 Tetracycline resistance
Although 59 different tetracycline resistance genes have been described (Marosevic et al., 2017), only the most frequent, tetM and tetO mediating resistance through ribosomal protection and tetK and tetL mediating resistance through efflux, were included in this study (Cho et al., 2020; Perreten et al., 2005). In the investigated enterococci isolates, tetracycline resistance was mainly characterized by the presence of tetM or a combination of tetM/tetL genes. Indeed, tetM was identified in 90.5% and 100% of the tetracycline-resistant E. faecalis (n = 48/53) and E. faecium (n = 65/65), respectively. A combination of tetM/tetL was reported in 79.2% of E. faecalis (n = 42/53) and 78.5% of E. faecium (n = 51/65), as tetL was always found in presence of tetM. tetO was isolated in 9 E. faecalis only, either alone (n = 5/9), in combination with tetM (n = 2/9), or with tetM/tetL (n = 2/9). tetK was not detected in enterococci in this study. In staphylococci, tetracycline resistance was reported in 98.4% of studied isolates (n = 61) and characterized by the presence of at least one tet gene, namely tetK (n = 2) and tetM (n = 7), or by a combination of them as following tetM/tetL (n = 12), tetM/tetK (n = 29), and tetM/tetL/tetK (n = 11). So, tetO was not detected among staphylococci. All tetracycline-resistant isolates from this study harbored at least one of the four targeted tet genes. More specifically, tetL was always found in combination with other tet genes, with tetM (n = 91) or tetM/tetO (n = 2) in enterococci, and tetM (n = 12) or tetM/tetK (n = 11) in staphylococci. In addition, tetL and tetM were found on the same contig in 19 of 25 (76.0%) sequenced enterococci, adjacent in 18 of them. In the latter, tetL and tetM were separated by ~3 kb but no insertion sequence (IS) was found between the two genes.
3.6 Oxazolidinone resistance
In this study, all linezolid-resistant (LZD-R) isolates (n = 22) collected through the Belgian official 2019–2020 monitoring of enterococci in food-producing animals were assayed. The cfr, optrA, and poxtA genes were targeted by this array. All linezolid-resistant isolates were harboring at least one linezolid-resistance gene: optrA (n = 14), poxtA (n = 2), or optrA/poxtA (n = 6). However, cfr was not detected among enterococci of this study, yet found in enterococci in Timmermans et al. (2022a). Linezolid resistance was not observed among the studied staphylococci isolates, and neither were optrA and poxtA detected. However, one linezolid-susceptible and chloramphenicol-resistant isolate characterized with an LZD MIC of 4 mg L−1 was harboring cfr, which presence was confirmed by WGS. Additionally, the presence of optrA and poxtA is not restricted to linezolid-resistant isolates since 9/15 enterococcal isolates exhibiting the LZD MIC of 4 mg L−1 (considered susceptible according to the ECOFF), were also harboring optrA (n = 6), poxtA (n = 2) or both (n = 1). Eight out of these nine isolates were resistant to chloramphenicol (MIC > 32 mg L−1) and one was susceptible to chloramphenicol (MIC = 32 mg L−1) as well. These genes were also found in enterococcal isolates characterized by a MIC of 2 mg L−1 (n = 5/60, 4 optrA, and 1 poxtA) and 1 mg L−1 (n = 2/27, 1 optrA, and 1 poxtA). Among these seven isolates, four were resistant to chloramphenicol (MIC > 32 mg.L−1) and three were susceptible to chloramphenicol (MIC = 16 mg L−1 [n = 2] and MIC = 32 mg L−1 [n = 1]).
3.7 Phenicol resistance
Among chloramphenicol-resistant-enterococci, genes coding for catpC194 (n = 2), catpC221-223 (n = 4), optrA (n = 1), poxtA (n = 2), optrA/catpC221-223 (n = 1), fexA/optrA (n = 19), optrA/poxtA (n = 3), and fexA/optrA/poxtA (n = 3) were detected. Surprisingly, catpC194 was detected in seven isolates phenotypically susceptible to chloramphenicol with MICs ranging from 16 mg L−1 (n = 1) to 32 mg L−1 (n = 6). Four enterococci harboring a fexA/optrA combination were characterized by a chloramphenicol MIC of 16 mg L−1 (n = 1) or 32 mg L−1 (n = 3). Similarly, optrA, poxtA, or a combination of both was found in 1, 4, and 1 chloramphenicol-susceptible isolates, respectively, characterized by MICs between 16 mg L−1 (n = 2) and 32 mg L−1 (n = 4). Among the 12 chloramphenicol-resistant staphylococci, nine carried the fexA gene, one carried cfr, while no relevant genes were detected in the two remaining isolates. Neither cat nor optrA or poxtA was detected in these chloramphenicol-resistant staphylococci. Three chloramphenicol-susceptible isolates harbored fexA and were characterized by a MIC of 8 mg L−1 (n = 2) or 16 mg L−1 (n = 1) for chloramphenicol. Besides the direct cross-resistance to linezolid and phenicols conferred by cfr, optrA, and poxtA, the concomitant presence of optrA and/or poxtA with the phenicol resistance gene fexA was observed in this study in 81.3% (n = 26/32) of the enterococcal isolates, as reported elsewhere (Brenciani et al., 2018; Ruiz-Ripa et al., 2020; Sadowy, 2018; Timmermans et al., 2022a). In addition, fexB was not included in the array but was detected in 10 sequenced enterococcal isolates, including five chloramphenicol-susceptible strains characterized by a MIC of 32 mg L−1. fexB was detected together with poxtA (n = 3), optrA/poxtA (n = 5), or fexA/optrA/poxtA (n = 2). Genome analysis indicated that the optrA/fexA combination was found on the same contig in 87.5% of isolates (n = 15/16). optrA and fexA were close to each other (~700 bp) in 14 of them and more distant in the remaining isolate (~5 kb). No IS was found between optrA and fexA in any of these isolates. The poxtA/fexB combination was never associated with the same contig, as described elsewhere (Freitas et al., 2020; Ruiz-Ripa et al., 2020; Timmermans et al., 2022a).
3.8 Glycopeptide resistance
Although rarely observed in 2019 and 2020 through the Belgian AMR monitoring program in enterococci, vancomycin resistance was however another point of interest of this study. Four probes targeting vanA, vanB, vanC1, and vanC2-3 genes were included in this array. vanA was detected in one of the two vancomycin-resistant E. faecalis isolates whereas vanB was not detected. Sequencing of the second isolate (VAR-660) revealed the unexpected presence of vanL. As expected, vanC1 and vanC2-3, intrinsic to E. gallinarum and E. casseliflavus, respectively, were not detected. Vancomycin resistance being absent in staphylococci from the studied years, no vancomycin-resistant isolates were tested with this array and in line with this, no van genes were detected with the array.
3.9 Aminoglycoside resistance
Many aminoglycoside resistance genes were described in the literature (Frye & Jackson, 2013; Hollenbeck & Rice, 2012; Schwarz et al., 2018; Strauss et al., 2015). The most common were selected and tested with this array, namely aadD, aadE, aacA-aphD, aphA3, and aph2-Id-Ie. In enterococci, gentamicin resistance was characterized by the presence of aacA-aphD in all resistant isolates (n = 37). Note, aph2-Id-Ie was not detected. Other genes conferring resistance to aminoglycosides not in the scope of the susceptibility testing (kanamycin and streptomycin) were also detected in enterococci, with aadD, aphA3, and aadE found in 2.4% (n = 3/124), 47.6% (n = 59/124), and 60.5% (n = 75/124) of all isolates, respectively. In staphylococci, susceptibility to gentamicin, kanamycin, and streptomycin was assessed by BMD testing. Kanamycin-resistant isolates (n = 19) were characterized by the presence of aacA-aphD/aadD (n = 12/19, 63.2%), aacA-aphD alone (n = 5/19, 26.3%) or aadD alone (n = 1/19, 5.3%). No aminoglycoside resistance gene could be detected in one kanamycin-resistant isolate. On the other hand, aadD was identified in seven kanamycin-susceptible isolates. Resistance to kanamycin was commonly associated with gentamicin resistance (n = 18/19) in accordance with the finding of gene combinations such as aacA-aphD (n = 5) or aacA-aphD/aadD (n = 12) in gentamicin-resistant isolates (n = 19). No aminoglycoside resistance gene targeted by the array could be detected in two gentamicin-resistant isolates. Resistance to streptomycin (n = 11) was correlated with the presence of aadE in four isolates. No aminoglycoside resistance gene could be detected with the array in the seven remaining streptomycin-resistant isolates. Neither aphA3 nor aph2-Id-Ie could be detected in any of the investigated aminoglycoside-resistant isolates.
3.10 Diaminopyrimidine resistance
Although susceptibility to trimethoprim was not assessed by BMD assays, dfr resistance genes were screened in enterococci. Among the 4 tested dfr genes, dfrD, dfrK, and dfrG were found in 1, 2, and 61 isolates, respectively. More precisely, dfrD was detected in 1 E. faecalis, dfrK in 2 E. faecium, and dfrG was detected in 35 E. faecalis and 26 E. faecium isolates. Also, dfrA/C was not detected in enterococci. Among the 55 trimethoprim-resistant staphylococcal isolates, the most previously reported dfr genes (Schwarz et al., 2018; Strauss et al., 2015; Wendlandt et al., 2013), namely dfrA/C, dfrD, dfrG, and dfrK, were targeted and found in 1, 0, 25 and 22 isolates, respectively.
3.11 Comparison of array-genotype with WGS-genotype: Short investigation
The AMR genetic profile of 31 isolates (16 E. faecalis, 14 E. faecium, and 1 MRSA) was investigated by whole-genome sequencing and compared to data resulting from the array. The selection of isolates for WGS was based on resistance to linezolid (n = 22), vancomycin (n = 2), or a minimum of 5 antimicrobials (n = 7). The genetic profiles gathered with the AMR array were all confirmed by WGS analyses (31/31) (see Table 5). These results ensured the reliability of the array method. Conversely, WGS investigation highlighted genes not covered by this array, that is vanL found in one vancomycin-resistant isolate (VAR-660).
| Isolate ID | Biosample ID | Species | Phenotypic resistance profile | Genetic resistance profile detected by the array and/or by WGS | Concordance ARRAY versus WGS (%) |
|---|---|---|---|---|---|
| VAR-660 | SAMN30360174 | Enterococcus faecalis | SYN VAN | lsaA, vanL2 | 100 |
| VAR-662 | SAMN30360175 | Enterococcus faecalis | CHL ERY GEN LZD SYN TET | aacA-aphD, aadE, aphA3, dfrG, ermB, fexA, lsaA, lsaE, lnuB, optrA, tetL, tetM | 100 |
| VAR-663 | SAMN30360176 | Enterococcus faecalis | CHL ERY GEN LZD SYN TET | aacA-aphD, aphA3, dfrG, ermA, ermB, fexA, lsaA, optrA, tetL, tetM | 100 |
| VAR-665 | SAMN30360177 | Enterococcus faecalis | CHL ERY LZD SYN | ermA, fexA, lsaA, optrA | 100 |
| VAR-668 | SAMN30360178 | Enterococcus faecalis | CHL CIP ERY GEN LZD SYN TET | aacA-aphD, aadE, aphA3, dfrG, ermB, fexA, lsaA, lsaE, lnuB, optrA, tetL, tetM | 100 |
| VAR-673 | SAMN30360179 | Enterococcus faecalis | CHL CIP ERY GEN SYN TET | aacA-aphD, aadE, aphA3, dfrG, ermB, fexA, lsaA, optrA, tetL, tetM | 100 |
| VAR-675 | SAMN30360180 | Enterococcus faecalis | CHL ERY LZD SYN TET | ermA, ermB, fexA, lsaA, optrA, tetL, tetM | 100 |
| VAR-676 | SAMN30360181 | Enterococcus faecalis | CHL ERY GEN LZD SYN TET | aacA-aphD, aadE, aphA3, ermA, ermB, fexA, lsaA, lsaE, lnuB, optrA, tetM | 100 |
| VAR-677 | SAMN30360182 | Enterococcus faecalis | CIP LZD SYN TET | dfrG, fexA, lsaA, optrA, tetL, tetM | 100 |
| VAR-678 | SAMN30360183 | Enterococcus faecalis | CHL ERY LZD SYN TET | dfrG, ermA, fexA, lsaA, optrA, tetL, tetM | 100 |
| VAR-682 | SAMN30360184 | Enterococcus faecalis | CHL ERY LZD SYN TET | ermB, fexA, lsaA, optrA, tetL, tetM | 100 |
| VAR-683 | SAMN30360185 | Enterococcus faecalis | ERY SYN TET VAN | aadD, aadE, aphA3, dfrD, lsaA, ermB, tetL, tetM, vanA, vatD | 100 |
| VAR-685 | SAMN30360186 | Enterococcus faecalis | SYN TET | lsaA, tetL, tetM | 100 |
| VAR-686 | SAMN30360187 | Enterococcus faecalis | CHL CIP ERY GEN SYN TET | aacA-aphD, aadE, aphA3, dfrG, ermA, ermB, fexA, lsaA, lsaE, lnuB, optrA, tetL, tetM | 100 |
| VAR-688 | SAMN30360188 | Enterococcus faecalis | ERY LZD SYN TET | aadE, aphA3, ermA, ermB, fexA, lsaA, optrA, tetL, tetM | 100 |
| VAR-689 | SAMN30360189 | Enterococcus faecalis | CHL ERY GEN LZD SYN TET | aacA-aphD, aadE, aphA3, dfrG, ermA, ermB, fexA, lsaA, lsaE, lnuB, optrA, tetL, tetM | 100 |
| VAR-661 | SAMN30360190 | Enterococcus faecium | AMP DAP ERY GEN SYN TET | aacA-aphD, aadE, aphA3, dfrG, ermA, ermB, lsaE, lnuB, tetL, tetM | 100 |
| VAR-664 | SAMN30360191 | Enterococcus faecium | AMP ERY GEN SYN TET | aacA-aphD1, aadE, aphA3, dfrG, ermB, lsaE, lnuB, poxtA, tetL, tetM | 100 |
| VAR-666 | SAMN30360192 | Enterococcus faecium | CHL ERY LZD TET | ermA, fexA, optrA, poxtA, tetL, tetM | 100 |
| VAR-667 | SAMN30360193 | Enterococcus faecium | CHL ERY GEN LZD SYN TET | aacA-aphD, aadE, aphA3, dfrG, ermA, ermB, lsaE, lnuB, optrA, poxtA, tetL, tetM | 100 |
| VAR-669 | SAMN30360194 | Enterococcus faecium | CHL LZD SYN TET | aadE, ermA, fexA, lsaE, lnuB, optrA, tetL, tetM | 100 |
| VAR-670 | SAMN30360195 | Enterococcus faecium | ERY GEN LZD SYN TET | aacA-aphD, aadE, aphA3, dfrG, ermA, ermB, lsaE, lnuB, optrA, poxtA, tetL, tetM | 100 |
| VAR-671 | SAMN30360196 | Enterococcus faecium | ERY LZD TET | dfrG, ermA, ermB, optrA, tetL, tetM | 100 |
| VAR-672 | SAMN30360197 | Enterococcus faecium | CHL ERY LZD TET | dfrG, ermA, optrA, poxtA, tetM | 100 |
| VAR-674 | SAMN30360198 | Enterococcus faecium | AMP DAP ERY GEN SYN TET | aacA-aphD, aphA3, aadE, catpC194, ermA, ermB, lsaE, lnuB, tetL, tetM | 100 |
| VAR-679 | SAMN30360199 | Enterococcus faecium | CHL ERY LZD SYN TET | aadE1, aphA3, dfrG, ermB, lsaE, lnuB, optrA, poxtA, tetL, tetM | 100 |
| VAR-680 | SAMN30360200 | Enterococcus faecium | LZD TET | aadD, poxtA, tetL, tetM | 100 |
| VAR-681 | SAMN30360201 | Enterococcus faecium | LZD SYN TET | poxtA, tetL, tetM | 100 |
| VAR-684 | SAMN30360202 | Enterococcus faecium | CHL LZD TET | fexA, optrA, poxtA, tetM | 100 |
| VAR-687 | SAMN30360203 | Enterococcus faecium | ERY LZD TET | dfrG, ermA, ermB, optrA, poxtA, tetL, tetM | 100 |
| VAR 659 | SAMN30360173 | Staphylococcus aureus | CHL CIP CLI ERY FOX PEN STR SYN TET TIA TMP | aadD, aadE, cfr, dfrG, ermB, lsaE, lnuB, tetK, tetL, tetM | 100 |
- Note: Genetic profiles include targets of this array and highlight consistency in comparison to WGS. Each gene detected by WGS was compared to the reference gene with a minimum percentage of coverage of 60% and a minimum percentage of identity of 90%.
- 1 Gene detected by WGS compared to the reference gene with a minimum percentage of coverage of 40% and a minimum percentage of identity of 40%.
- 2 Gene detected by WGS, not targeted by the ARRAY.
4 DISCUSSION
Among the entire collection of 62 staphylococci and 124 enterococci investigated, a total of 91.3% (n = 718/786) resistance profiles demonstrated experimentally could be associated with the AMR gene targeted by the array, namely 89.2% (n = 305/342) in staphylococci and 93.0% (n = 413/444) in enterococci. This result supports that the most common resistance genes were targeted by our array since a genetic marker could be associated with more than 90.0% of the resistant phenotypes leaving only a few without genetic information.
This study highlighted the coexistence of both lsaA and lsaE in 20/57 (35.1%) of the E. faecalis isolates, suggesting the presence of a selection pressure since lsa genes are involved in other phenotypic resistances such as pleuromutilin (tiamulin) and lincosamide (clindamycin) resistance as reported in other studies (Feßler et al., 2018; Hollenbeck & Rice, 2012). However, antimicrobial susceptibility for these antimicrobial classes is not monitored in enterococci in official surveillance programs. In addition, lsaE was the most observed gene in MRSA with 29 isolates showing resistance to Q/D (n = 29/37; 78.4%), as well as to tiamulin (n = 29/35; 88.6%) and clindamycin (n = 29/57; 50.9%). Such pleuromutilin-lincosamide-streptogramin A resistance mediated by lsaE as found in our study among Staphylococcus spp. and Enterococcus spp. isolated from animals was reported elsewhere in animals and also in humans (Feßler et al., 2018; Schwarz et al., 2018; Wendlandt et al., 2013).
Overall, ermB was the most prevalent macrolide resistance gene in our collection (in 79.6% of all erythromycin-resistant isolates, n = 113/142) and particularly among enterococci (n = 98/106 vs. n = 15/36 staphylococci), as described in Frye & Jackson (2013). The erm genes were more frequently observed in enterococci than staphylococci (97.2% vs. 77.8%) as first described in Jensen et al. (1999). Interestingly, other studies (Jensen et al., 1999; Petinaki & Papagiannitsis, 2019) also reported ermC mostly in staphylococci and in rare cases in enterococci.
lnuB was the second most observed lincosamide resistance gene in staphylococci as found in 45.6% of isolates (n = 26), however, it has been mostly described in enterococci (Cho et al., 2020; Feßler et al., 2018) and rare cases in staphylococci (Li et al., 2013; Lozano et al., 2012; Wendlandt et al., 2013).
All tetracycline-resistant isolates of this study harbored at least one of the tested genes, namely tetL, tetM, tetK, and/or tetO, with tetM being the most prevalent one as reported elsewhere (Argudin et al., 2017; Cho et al., 2020; Frye & Jackson, 2013; Schwarz et al., 2018). So, tetL was always found in combination with other tet genes, particularly with tetM which was adjacent in 72.0% (n = 18/25) of the isolates investigated by WGS. This result suggests that tetM and tetL might be transferable together thereby spreading two different mechanisms of resistance, ribosomal protection, and proteins efflux, respectively.
In this study, at least one linezolid-resistance gene was detected in each LZD-R strain: optrA, poxtA, or a combination of both genes in 63.6%, 9.1%, and 27.3% of LZD-R enterococcal isolates, respectively. These genes were also found in LZD-susceptible isolates characterized by a MIC of 4 mg L-1 (n = 9/15), 2 mg L-1 (n = 5/60), or 1 mg L-1 (n = 2/27). These results support a recent warning from EFSA suggesting that all strains displaying MICs ≥ 4 mg L-1 for linezolid and exhibiting resistance to the other compounds typically conferred by cfr, should be screened for this gene (European Food Safety Authority & European Centre for Disease Prevention and Control, 2022, p. 116). Perhaps it should be considered to extend it to optrA and poxtA screening as well, especially as they have been mostly found in enterococci (Timmermans et al., 2022a). In line with the observation of optrA or poxtA in LZD-susceptible isolates, previous studies (Dejoies et al., 2020; Timmermans et al., 2022a) demonstrated that a 48 h-incubation conducted during susceptibility testing enhanced the detection of isolates carrying linezolid-resistance determinants suggesting that optrA and poxtA might be inducible. However, more experiments are required at this point. Few chloramphenicol susceptible isolates were also characterized by the presence of a resistance gene such as fexA (n = 4) and catpC194 (n = 7) as described for LZD-susceptible isolates here above. catpC194, catpC221, and catpC223 from the cat enzyme family as well as fex exporters are inducible (Schwarz et al., 2016), which could explain the variations observed among MICs. This highlights the limitations of AST based on phenotypic cut-offs for the screening of AMR genes, in particular for such inducible genes. In addition, future genetic investigation of chloramphenicol phenotypic resistance may partially rely on the detection of fexB, a member of the fex exporters family already reported in enterococci (Argudin et al., 2017; Schwarz et al., 2018) and found in 10 sequenced enterococci of this study. Interestingly, the concomitant presence of optrA and fexA was observed in 23 of our isolates independently of phenotypic resistance profiles. Particularly, optrA was found close to fexA in 14 of the 16 isolates investigated by WGS. The absence of IS between the two genes in these isolates suggests both genes might be transferable together.
Vancomycin resistance, although rare, was characterized by the presence of the vanA cluster in one of the two isolates, the most common gene reported in other studies (Courvalin, 2006; Ekwanzala et al., 2020; Torres et al., 2018). In the remaining vancomycin-resistant isolate (VAR-660), sequencing highlighted the presence of the rare vanL, a gene not covered by this array. The rare vanL gene cluster, first described by Boyd et al. (2008), has been so far detected on the chromosome of a single E. faecalis isolate of human origin (Ekwanzala et al., 2020) displaying low-level vancomycin resistance (Ekwanzala et al., 2020; Boyd et al., 2008) as observed here (MIC = 8 mg L−1). Our study reports the presence of vanL in an E. faecalis isolate from animal origin. The origin of the vanL gene cluster and the way an animal (i.e., a pig) acquired this strain remain elusive.
Gentamicin resistance was characterized by the presence of aacA-aphD in all resistant enterococci (n = 37) and by aacA-aphD (n = 5) or aacA-aphD/aadD (n = 12) in resistant staphylococci (n = 19) of this study. In addition, resistance to gentamicin was found to be associated with kanamycin resistance in 18 of these 19 resistant staphylococci, with kanamycin-resistant isolates (n = 19) harboring aadD alone (n = 1), aacA-aphD (n = 5) or aacA-aphD/aadD (n = 12) gene combination.
In a few cases of this study (10.8% of the individual resistant phenotypes in staphylococci and 7.0% in enterococci), a genetic marker could not be associated with the resistant phenotype. In enterococci, no relevant AMR genes were found with the array to explain erythromycin (n = 3), Q/D (n = 6), chloramphenicol (n = 10), gentamicin (n = 1) and vancomycin (n = 1) phenotypic resistances (out of a total of 444 assessed phenotypes). In staphylococci, a genetic explanation was missing for clindamycin (n = 1), erythromycin (n = 6), Q/D (n = 2), tiamulin (n = 6), chloramphenicol (n = 2), gentamicin (n = 2), streptomycin (n = 7), and trimethoprim (n = 7) resistant phenotypes (out of a total of 342 assessed phenotypes). In addition, the results of this study highlighted the complexity of Q/D resistance and a lack of a complete genetic explanation in 14/90 isolates since the identified genes (except for lsaA and lsaE) have been reported to confer resistance to either streptogramin A or streptogramin B but not both. Particularly, the presence of erm (streptogramin B) alone in Q/D-resistant isolates (10/53 E. faecium, 4/37 MRSA) does not explain the observed Q/D-resistant phenotype. Similar to lsa, eatA would be an interesting target as this gene confers the same profile of cross-resistance to lincosamides, streptogramins A, and pleuromutilins (Isnard et al., 2013). The WGS investigation revealed the presence of msrC in one E. faecium (VAR-681) exhibiting resistance to Q/D, this gene being frequently reported elsewhere in this species (Frye & Jackson, 2013; Hollenbeck & Rice, 2012). And, msrC has been described in Q/D-susceptible isolates, however first reported as being specifically found in resistant E. faecium. This suggests that it might be silent or involved in resistance to other antibiotics (Smoglica et al., 2022) such as macrolides (Portillo et al., 2000; Zaheer et al., 2020). Therefore, msrA which shares significant sequence identity with msrC has been reported to confer macrolides-streptogramin B resistance in staphylococci and could be an interesting future target to investigate erythromycin and/or Q/D phenotypic resistances (Reynolds & Cove, 2005). In addition, ermF, ermY, mphB or more recently described mefD, msrF, and msrH have been found in staphylococci to confer macrolide and lincosamide resistances (Miklasinska-Majdanik, 2021; Schwarz et al., 2018; Woodford, 2005). Besides, ereA and ereB esterases have been reported to confer macrolide resistance in staphylococci isolated from animals as well (Miklasinska-Majdanik, 2021; Schwarz et al., 2018). In addition, chromosomal mutations in rplD or rplV coding for ribosomal proteins L4 and L22 leading macrolide and Q/D resistance, respectively, were also observed in Streptococcus pneumoniae and S. aureus and could be targeted in future studies (Farrell et al., 2004; Malbruny et al., 2002; Miklasinska-Majdanik, 2021). The sal gene family reported to confer lincosamide, streptogramin A and pleuromutilin resistance in staphylococci from humans or animals at various levels could be investigated in an attempt to explain the tiamulin-resistant phenotypes observed in 6/35 isolates (Mohamad et al., 2022; Schwarz et al., 2018). Pleuromutilin resistance also derives from chromosomal mutations in 23S rRNA or rplC as described in staphylococci isolated from both humans and animals (Paukner & Riedl, 2017; Van Duijkeren et al., 2014). No gene of this array was detected in 10 isolates (1 E. faecalis and 9 MRSA) exhibiting resistance to aminoglycosides, namely to gentamicin (n = 2 MRSA and 1 E. faecalis) and to streptomycin (n = 7 MRSA). In enterococci, an intrinsic low-level of aminoglycoside resistance can be occasionally observed due to the presence of the species-specific chromosomal aac(6')-Ii gene found in almost all E. faecium (Adamecz et al., 2021). Among many aminoglycosides modifying enzymes (AMEs) described in the literature, a few of them such as str could be interesting to target in the future as already described in staphylococci (Ramirez & Tolmasky, 2010; Schwarz et al., 2018). In contrast, few cases of mismatch were observed with kanamycin-susceptible isolates (n = 7 MRSA) by BMD testing carrying one gene encoding aminoglycosides resistance (aadD in this study), as already reported in other studies (Adamecz et al., 2021; Feizabadi et al., 2006; Yean et al., 2007). This could be explained if this gene is nonfunctional and could be investigated in future studies. In addition, Ida et al. (2002) have shown that rearrangements caused by the integration of insertion elements into the staphylococcal chromosome or plasmids affected the expression of adjacent genes including aminoglycoside resistance genes. Finally, 7/55 (12.7%) trimethoprim-resistant MRSA isolates were not found to be associated with the presence of a genetic marker in this study. The presence of other members of the dfr genes family (e.g., dfrF) or the presence of chromosomal mutations may explain the phenotypic resistance observed in these isolates (Woodford, 2005).
5 CONCLUSIONS
In this study, a bead array was described aiming to detect broad AMR genetic profiles of Gram-positive bacteria from healthy animals in a single experiment. A number of resistance genes circulating among Enterococcus spp. and Staphylococcus spp. were targeted by this array allowing the screening of a large number of strains in a limited time and at an affordable cost. The relatively short turnaround time (~8 h) and the use of a software source code freely available allow rapid data acquisition and analysis. In addition, the method is cost-effective with a reagent price of 18 €/sample. The designed array targeted AMR from 9 antimicrobial families, including resistance to critically important antimicrobials, linezolid, and vancomycin, which are important to monitor in both human and animal sectors. Due to the flexibility of the method, a more specific bead array (e.g., linezolid-array) could also be easily designed by removing/adding probes to respond to a particular epidemiologic situation (i.e., location) or to screen other bacterial species; and could also be extended to other research areas (i.e., virulence). Animal isolates were investigated; nevertheless, other reservoirs of AMR genes (humans, environment) could be assessed with the array. Besides AMR genes, resistance resulting from point mutations in receptors (e.g., ciprofloxacin resistance) or in specific proteins (e.g., daptomycin resistance) are targetable as well. In parallel to the official monitoring based on AST, screening with the array allowed us to rapidly determine genetic profiles circulating among livestock animals in Belgium. Isolates of this study have frequently been shown to carry two or more resistance genes conferring the same resistance phenotype, and sometimes genes from the same family, for example erm or tet genes. This accumulation of genes has been frequently observed while a single gene is sufficient to confer resistance, and may be explained by acquisition at different times and/or under different conditions, including their possible co-location on plasmids carrying multiple resistance genes even in absence of selective pressure (Schwarz et al., 2018). Many studies suggested that commensal bacteria are probable reservoirs for AMR genes and can transfer these to pathogenic organisms, for example Bacillus spp. or Salmonella spp. (Frye & Jackson, 2013; Schwarz et al., 2018). Also, while the spread of resistance genes such as ermB and tetM is limited to Gram-positive bacteria, genes such as aadE or tetL are found both in Gram-positive such as reported here but also in Gram-negative bacteria (E. coli) as reported elsewhere (Frye & Jackson, 2013; Schwarz et al., 2018). This array allowed the identification of AMR genes in resistant isolates, spotting a few isolates without genetic explanation as interesting candidates for WGS to identify new resistance mechanisms. This approach allowed us to demonstrate the presence of vanL in one E. faecalis isolate from animal origin. Our study also highlighted the presence of the lnuB gene in staphylococci, rarely reported in the literature. Finally, the missing concordance between aminoglycoside resistance genes and phenotypic resistance observed in this study illustrates the importance of still relying on the routine phenotypic susceptibility test for resistance monitoring. Oppositely, the presence of an oxazolidinone resistance gene in a susceptible isolate occurred several times and showed the importance of genotyping as a complement to phenotyping, particularly for such potentially inducible genes. Our data stress the importance of interpreting AMR with caution and the complementarity of both phenotyping and genotyping methods.
AUTHOR CONTRIBUTIONS
Carole Kowalewicz: Conceptualization (lead); formal analysis (lead); methodology (lead); writing – original draft (lead); writing – review and editing (equal). Michael Timmermans: Conceptualization (supporting); methodology (supporting); writing – review and editing (supporting). David Fretin: Writing – review and editing (supporting). Pierre Wattiau: Conceptualization (supporting); supervision (supporting); writing – review and editing (supporting). Cécile Boland: Conceptualization (supporting); formal analysis (supporting); methodology (supporting); supervision (lead); writing – original draft (supporting); writing – review and editing (equal).
ACKNOWLEDGMENTS
We thank the Federal Agency for the Safety of the Food Chain for collecting the samples and isolates used in this study. We thank A. Radu for technical support, technicians of the service Transversal Activities in Applied Genomics at Sciensano, Belgium for performing Next Generation Sequencing runs and the development and maintenance of the in-house instance of the Galaxy workflow management system. We also thank V. Perreten, A. J. O'Neill, S. Schwarz, F. Fux of the Bacterial Diseases Unit at Sciensano, and the EURL-AR for providing strains used for the array validation.
CONFLICT OF INTEREST
None declared.
ETHICS STATEMENT
None required.
APPENDIX
| Species | Strain ID | Gene | Source |
|---|---|---|---|
| Enterococcus faecalis | VAR-473 | cfr | Timmermans et al. (2022a) |
| Enterococcus faecalis | VAR-473 | poxtA | Timmermans et al. (2022a) |
| Enterococcus faecalis | VAR-473 | optrA | Timmermans et al. (2022a) |
| Enterococcus faecium | VAR-182 | fexA | In-house collection |
| Enterococcus spp. | VAR-181 | vanA | In-house collection |
| Enterococcus faecalis | V583 | vanB | EURL-AR |
| Enterococcus gallinarum | VAR-530 | vanC1 | Timmermans et al. (2022a) |
| Enterococcus casseliflavus | VAR-484 | vanC2-3 | Timmermans et al. (2022a) |
| Enterococcus faecium | VAR-172 | catp C194 | In-house collection |
| Enterococcus spp. | VAR-181 | catp C221-223 | In-house collection |
| Enterococcus casseliflavus | UC 73 Id | aph2-Id/Ie | EURL-AR |
| Staphylococcus aureus | VAR-141 | aacA-aphD | In-house collection |
| Enterococcus faecium | VAR-182 | aphA3 | In-house collection |
| Staphylococcus aureus | VAR-141 | aadD | In-house collection |
| Enterococcus faecium | VAR-182 | aadE | In-house collection |
| Enterococcus faecium | S. A plasmid pG01 | dfrA/C | Caryl & O'Neill (2009) |
| Enterococcus spp. | VAR-181 | dfrD | In-house collection |
| Enterococcus faecium | VAR-182 | dfrG | In-house collection |
| Snterococcus aureus | VAR-134 | dfrK | In-house collection |
| Streptococcus spp. | 01D19 | mefA | Internal Sciensano collection |
| Streptococcus spp. | 02J1175 | mefE | Internal Sciensano collection |
| Enterococcus faecium | VAR-182 | ermA | In-house collection |
| Enterococcus faecium | VAR-182 | ermB | In-house collection |
| Bacillus subtilis | B.3HU104/pE194 | ermC | EURL-AR |
| Staphylococcus chromogenes | TS1 - NC_007768.1 | lnuA | Luthje et al. (2007) |
| Enterococcus faecium | VAR-182 | lnuB | In-house collection |
| Enterococcus faecalis | VAR-175 | lsaA | In-house collection |
| Enterococcus faecium | VAR-182 | lsaE | In-house collection |
| Staphylococcus aureus | BM 3093, pIP680 | vatA | EURL-AR |
| Staphylococcus spp. | 9674438-1vatB-pos. | vatB | EURL-AR |
| Staphylococcus cohnni | BM10711, pIP1675 | vatC | EURL-AR |
| Enterococcus faecium | BM4145 | vatD | EURL-AR |
| Enterococcus faecium | UW1965 | vatE | EURL-AR |
| Staphylococcus aureus | VAR-134 | vgaA | In-house collection |
| Staphylococcus aureus | BM12235, pIP1633 | vgaB | EURL-AR |
| Staphylococcus cohnni | BM10711, pIP1675 | vgbB | EURL-AR |
| Staphylococcus xylosus | LT223129.1 (JW2311) | mphC | Wipf et al. (2017) |
| Enterococcus faecium | VAR-182 | tetM | In-house collection |
| Enterococcus faecium | VAR-182 | tetL | In-house collection |
| Staphylococcus aureus | VAR-143 | tetK | In-house collection |
| Enterococcus faecalis | VAR-175 | tetO | In-house collection |
| Staphylococcus aureus | ATCC 29213 | staph | ATCC collection |
| Enterococcus faecalis | ATCC 29212 | sodA-fs | ATCC collection |
| Enterococcus faecium | E. faecium 2013/16227 | sodA-fm | In-house collection |
- Note: Positive control of vgaD was absent in this study.
- Strains with source as “In-house collection” were isolates sequenced by whole-genome sequencing of our collection.
Open Research
DATA AVAILABILITY STATEMENT
All data analyzed during this study are included in this published article. The WGS datasets are available under BioProject PRJNA868571: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA868571.