Deciphering the role of host species for two Mycobacterium bovis genotypes from the European 3 clonal complex circulation within a cattle-badger-wild boar multihost system
Graphical Abstract
Using whole genome sequencing of Mycobacterium bovis strains collected in two distinct areas of France with an evolutionary model, we showed that the role of host species in the circulation of the pathogen differed between both areas. While wild boars appeared to play the role of an intermediary between badgers and cattle in both areas, the role of badgers differed. Our results suggest that the transition pattern depends on ecological, landscape, and anthropic factors.
Abstract
Bovine tuberculosis is a common disease affecting cattle and wildlife worldwide. Mycobacterium bovis circulation in wildlife decreases the efficacy of surveillance and control programs in cattle. Strains of the European 3 clonal complex are the most frequent in France. The aim of our work was hence to investigate the role played by cattle and wildlife species in the circulation of two M. bovis European 3 strains circulation. WGS of M. bovis strains collected between 2010 and 2017 in two distinct areas (Nouvelle-Aquitaine region, NAq, and Côte-d'Or département, CdO), from badgers, wild boars, and cattle were used in an evolutionary model to infer the transition between the three species. We computed host species transition and persistence between two consecutive nodes and the average number of transitions per tree. In total, 144 and 218 samples were collected respectively in CdO and NAq. In CdO, three between-species transition rates stood out: from cattle to badgers, from badgers to wild boars, and from wild boars to cattle. In NAq an additional fourth transition rate was identified: from badgers to cattle. However, host transition remained a rare event. Our results suggest that wild boars could be an intermediary host between badgers and cattle in the circulation of the studied strains in CdO and NAq. Our results also highlight the differences between these two areas, suggesting that the transition pattern does not only depend on the host species and other ecological, landscape and anthropic factors are important.
1 INTRODUCTION
Bovine tuberculosis (bTB) is a disease affecting cattle and wildlife worldwide (Bovine tuberculosis (2021)). Mycobacterium bovis can infect a large variety of wildlife hosts (Fitzgerald & Kaneene, 2013), which differ from country to country. M. bovis was detected in European badgers (Meles meles) in the UK (Rivière et al., 2014), Ireland, and continental Europe, in wild boars (Sus scrofa) in continental Europe (Rivière et al., 2014), in red foxes (Vulpes vulpes) in France (Michelet et al., 2018), in cervids, more specifically red deer (Cervus elaphus) and roe deer (Capreolus capreolus) in continental Europe (Rivière et al., 2014), Sika deer (Cervus nippon) recently identified in Ireland (Kelly et al., 2021) and white-tailed deer (Odocoileus virginianus) and elk (Cervus canadensis) in North America or brush-tailed possums (Trichosurus vulpecula) in New Zealand (Fitzgerald & Kaneene, 2013). The circulation of M. bovis in wildlife hampers control programs when implemented.
In France, a control program in line with the European Union (EU) Directive 64/432/EEC has been implemented starting in 1954 to eradicate bTB in cattle farms. This program led to a rapid decrease in herd incidence (Michelet et al., 2020), resulting in a disease-free status in 2001 when herd prevalence was below 0.1% for six consecutive years (Decision 2001/26/EC). The bTB-free status alleviates the control measures for export and therefore ensures French cattle farming competitiveness.
There are two surveillance programs in France: one for cattle and one for wildlife. The cattle surveillance program relies on three components. First, at slaughterhouses at the national level, all carcasses are systematically inspected, with incisions of specific tissues (lungs, retropharyngeal, tracheobronchial, and mediastinal lymph nodes). Samples from suspect lesions are sent for bTB confirmation by polymerase chain reaction (PCR) or bacteriology to certified laboratories. Second, depending on the epidemiological situation in the département (intermediate administrative divisions of France), periodic systematic screening of all animals over 6 weeks of age is performed at a regularity ranging from yearly to none. Finally, based on the epidemiological investigation of bTB-infected farms, targeted screening is performed on animals before they depart from at-risk farms (Delavenne et al., 2020a).
The wildlife surveillance program created in 2011, called Sylvatub, focuses on red deer, roe deer, wild boars, and badgers (Rivière et al., 2014). It is based on event-based surveillance (e.g. carcass inspection of hunted wild boars and cervids), enhanced event-based surveillance (e.g., carcass inspection of badgers found dead on the roadside), and programmed surveillance (e.g. badger trapping or hunted wild boar direct diagnosis). The implementation of these modalities depends on the level of surveillance defined at the département level, which in turn depends on their epidemiological situation. Animals are necropsied and appropriate samples are taken for bTB diagnosis by PCR and culture and molecular typing when confirmed positive (Réveillaud et al., 2018).
In France, the epidemiological situation is heterogeneous. Few areas concentrate most national outbreaks, such as Côte-d'Or département in the East of France or the Nouvelle-Aquitaine region in the South-West. As a result, in these two areas, surveillance of cattle was biennial until 2018 and is annual since 2018 in the Dordogne département; and for wildlife, the program includes event-based, enhanced event-based, and programmed surveillance (Delavenne et al., 2020b).
In addition, molecular typing revealed that M. bovis strains circulating in these two regions are specific per area. For instance, SB0120 VNTR profile 5 3 5 3 9 4 5 6 (SB0120-NAq) is mainly found in an area of Nouvelle-Aquitaine overlapping Dordogne, Haute-Vienne, Charente and Charente-Maritime départements, while SB0120 VNTR profile 5 5 4 3 11 4 5 6 (SB0120-CdO) is mainly found in Burgundy, especially Côte-d'Or département (Michelet et al., 2020). To differentiate strains of these dominant genotypes within these regions at a finer scale, a more discriminating method is required such as whole genome sequencing (WGS), which has shown higher resolution (Crispell et al., 2017; Price-Carter et al., 2018).
Despite being phylogenetically close genotypes belonging to the European 3 (Eu3) clonal complex (also described as lineage La1.2 (Zwyer et al., 2021)) affecting the same host species (Hauer et al., 2015, 2019), the epidemiologic situations were contrasted in these two areas. While the number of cattle outbreaks has been steadily decreasing in CdO, the number of detected cases has been increasing in NAq (Delavenne et al., 2020a). In wildlife, a similar trend was observed in badgers, with the apparent prevalence decreasing from 8.1% to 4.2% in Côte-d'Or between 2013 and 2014 (as identified by culture) and 2016–2017 (as identified by PCR) while it increased in Nouvelle-Aquitaine from 2.7% to 5.3% (Réveillaud et al., 2018). However, the apparent prevalence of wild boars decreased in both areas (from 3.1% to 2.2% in Côte-d'Or and from 4.1% to 2.7% in Nouvelle-Aquitaine during the same time intervals) (Réveillaud et al., 2018).
The drivers of these epidemic dynamics are still unclear. In particular, the role played by each species in these two multi-host systems remains to be determined. Previous studies on bTB have focused on interactions between two species: primarily cattle and badgers (Biek et al., 2012; Bouchez-Zacria et al., 2018; Crispell et al., 2019; Rossi et al., 2020; Trewby et al., 2014) but also cattle and possums (Crispell et al., 2017), cattle and elk (Salvador et al., 2019), cattle and cervids (Crispell et al., 2020), or wild boars and cervids (Zanella et al., 2008). However, species such as badgers and wild boars have different life traits: while badgers are sedentary and have a life expectancy of about 14 years, wild boars can travel long distances and are often hunted before they are 4 or 5 years old (Byrne et al., 2014; Podgórski et al., 2013). In addition, the amount and the timing of M. bovis shedding by the different species, hence their infectiousness, may differ. Thus, the roles played by different species in the multi-host system could be different.
The aim of our work was therefore to investigate the role played by each species in the circulation of two Eu3 M. bovis lineages using genomic data. To do so, we defined two study areas in CdO and NAq, in which samples collected from cattle, badgers, and wild boars have been analyzed by WGS. We then used these data to model the evolutionary history of the pathogen and to infer host species of ancestors. This allowed us to analyze the transitions between species in a three-species multi-host system.
2 MATERIAL AND METHODS
2.1 bTB detection
All bTB detections which have been declared to the Directorate General on Food Safety (Direction Générale de l'Alimentation – DGAl) between 2010 and 2017 included herd identification for cattle and date of bTB confirmation. The National Reference Laboratory (NRL) (Anses, Maisons-Alfort) verified the data set for consistency with the samples that are analyzed at the lab for bTB confirmation.
Suspect cattle had been identified either by skin tests (in the cervical region, using single intradermal tuberculin test (SITT) or single intradermal comparative tuberculin test (SICTT)) provided by different surveillance protocols (periodic screening, epidemiological investigations, pre-movement of cattle (Delavenne et al., 2020a)) or following the detection of lesions at the slaughterhouse. For each detection, the presence of M. bovis was confirmed by PCR and/or bacterial culture (Delavenne et al., 2020a). Isolated strains are genotyped by spoligotyping and MIRU-VNTR. Only isolates belonging to the SB0120 spoligotype were included, more specifically isolates of genotype SB0120-CdO in CdO and isolates of genotype SB0120-NAq in NAq.
Wildlife animals shot during hunting or found dead were subject to a necropsy and samples were collected to detect mycobacteria by PCR and culture. All strains isolated from badgers and wild boars identified during the wildlife surveillance program in the study area were included in our study. We decided to exclude samples collected from red deer, roe deer, and red foxes since their limited number (n = 2, 1, and 5, respectively) would have altered parameter inference (Réveillaud, 2013).
2.2 Study areas selection
In CdO and NAq, study areas were defined according to the following criteria: (1) the municipality with the most isolates was included; (2) the final study area was well delimited (i.e., it was mostly surrounded by municipalities without detected cases); (3) the final study area was compact (i.e., with a limited number of municipalities without cases detected within it). The municipalities with detected cases were municipalities where infected wildlife was identified or municipalities with pastures belonging to farms with outbreaks. The pastures were identified with the 2013–2018 French graphic parcel register (Relevé Parcellaire Graphique, RPG) provided by the French Ministry of Agriculture. We call below “outbreak” the official declaration of one or several bTB-infected animals in a given farm. A farm could hence have several outbreaks (in case of breakdown after the farm has recovered the bTB-free status). We limited the number of samples to be analyzed per outbreak to three. Indeed, in NAq, the number of available isolates per outbreak varied between one and 26 and was <3 in most outbreaks (84%). As two or three different VNTR profiles were identified in some outbreaks (2.5%), bounding the number of isolates per outbreak to three allowed limiting the number of isolates to sequences, while allowing for the detection of different sequences in the same outbreak.
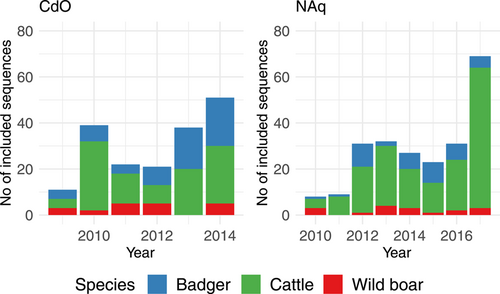
CdO was the pilot area and the municipalities were chosen manually according to the detection of infected wildlife or localization of pastures belonging to infected farms. In CdO, the selected area consisted of 38 municipalities, covering 499 km², in which 144 isolates (only one M. bovis strain per individual animal) were selected between 2009 and 2014. The number of samples to be analyzed was limited to three per outbreak. The host species were cattle (n = 77, from 74 outbreaks), badgers (n = 52) and wild boars (n = 15) (Figure 1, left panel) (Table A1).
For NAq, we decided to formalize the municipality selection process with a procedure that reflects the decisions taken to select the municipalities in CdO. First, for each municipality, the number of samples available was defined as the total of available samples collected from the outbreak and the wildlife. We then defined the “starting zone” as the municipality with the most available samples and the neighboring municipalities. We iteratively added one municipality randomly selected from the list of neighboring municipalities with at least one sample. The added municipality was thus adjacent to the previously defined area. Iterations stopped once a predefined maximal number of samples to include (nmax) was reached. We ran this iterative process 1000 times. Among the 1000 areas thus obtained, we selected the best-delineated area, minimizing the ratio of the total length of borders with noninfected municipalities to the perimeter of the selected area.
The resulting selected area in NAq consisted of 76 municipalities, covering 1540 km², in which nmax = 219 samples collected between 2010 and 2017 were selected. The host species were cattle (n = 161, from 95 outbreaks), badgers (n = 41) and wild boars (n = 17) (Figure 1, right panel). In CdO, the maximum number of samples was collected in 2014 (n = 51), the minimal number in 2009 (n = 11) with no clear trend, whereas in NAq, the number of selected samples varied between 8 in 2010 and 69 in 2017 with a trend toward an increase with time (Figure 1).
2.3 Sequencing and alignment
Thermolysates of selected isolates were sequenced by Illumina sequencing (paired-end 2*250 bp) at Genoscreen (Pasteur Institute, Lille) for CdO and Illumina sequencing (paired-end 2*150 bp) at the Paris Brain Institute (ICM) for NAq. Sequencing quality was controlled using FASTQC with an acceptability Phred score threshold of 30. Sequence alignment and Single-nucleotide polymorphism (SNP) calling were computed at the NRL using the Mb3601 reference strain (Branger et al., 2020) on Bionumerics software, version 7.6 (AppliedMath, Belgium). Identified SNPs were selected according to strict criteria of the wgSNP module: (1) they had to be present on at least five reads in both forward and reverse direction, (2) 12 base pairs had to separate them, (3) they were not present in repetitive regions of the genome, and (4) ambiguous SNPs (at least one unreliable (N) base, ambiguous (non ATCG) base or gap) were not included. SNPs were then used to reconstruct a maximum parsimony tree on Bionumerics to identify genetic outliers.
2.4 Bayesian phylogenetic modeling
We used BEAST2 (Bayesian evolutionary analysis by sampling trees) 2.6.4 to model bTB evolutionary (Bouckaert et al., 2019). The sequences for each region were analyzed separately.
2.5 The structured coalescent model
The differences in surveillance protocols between cattle and wildlife induce sampling biases. Indeed, wildlife cannot be exhaustively monitored, while all slaughtered cattle are tested for bTB. To take into account this sampling bias, we used the approximation of the structured coalescent as implemented in the Mascot (Marginal Approximation of the Structured COalescenT) 2.1.2 package (Müller et al., 2018). Indeed structured coalescent population models, contrary to migration models, are less susceptible to sampling bias (Müller et al., 2017). We assumed constant between-species transition rates in times and used the Bayesian stochastic search variable selection (BSSVS) procedure to select only transition rates that explain the transition of M. bovis between the different species (Lemey et al., 2009). This procedure was designed to limit the number of transition rates to infer only those adequately explaining the diffusion between the subpopulations (here the host species) (Lemey et al., 2009). The estimated transition rates were backward in time, however, to avoid confusion we present the transition rates as the forward transition rates thereafter.
2.6 Substitution and molecular clock model selection
To select the best-fitting model, the marginal likelihood (ML) was computed using the nested sampling algorithm as implemented in the NS 1.1.0 package (Russel et al., 2019). Models were then compared two by two by computing the Bayes factor (BF) as BF = log(ML2)-log(ML1), where ML1 and ML2 are the ML of models 1 and 2, respectively. The level of support was considered overwhelming when |BF | > 150, strong if 20 < |BF | ≤ 150, positive 3 < |BF | ≤ 20 and hardly worth mentioning if 1≤|BF | ≤ 3 (Kass & Raftery, 1995). The model with the largest ML was favored. We tested the substitution models (JC69, HKY, and GTR) and three molecular clock models (strict, uncorrelated exponential, and uncorrelated lognormal relaxed molecular clock).
2.7 Parameters inference
To infer the parameters, we set the Markov chain Monte Carlo (MCMC) length to 50 million, the burn-in to 10%, and sampled every 5000 iterations. Four replicates were performed. For each replicate, we inspected each parameter trace, looking for a stationary distribution, as well as the effective sample size (ESS). We considered ESS > 200 would guarantee that samples were independent and only selected models for each of the inferred parameters. We combined the four replicates with LogCombiner v2.6.6 with a lower sampling frequency to analyze thereafter 10,000 parameters. All trees were plotted using the ggtree package in R (Yu et al., 2017).
2.8 Host species transition
To study host species transitions, we resampled 1001 trees. We recorded for each node of each tree the predicted host species, its probability, and its height. We considered that the host species was known if its probability was >0.9 and unknown otherwise. We then computed the number of host species transitions between two consecutive nodes as well as the number of host species persistence (i.e., when the descendant host species is the same as the parental host species). If the host species for the parent and/or the descendant node was unknown, then the transition was considered unknown as well. We computed the host species transition and persistence at the lineage level. It is noteworthy that two consecutive nodes do not necessarily represent two distinct hosts; indeed these two nodes could represent strains hosted by the same individual or conversely strains that could have been transmitted to one or several other individuals. We computed the average number of transitions per tree for each parental node time as the sum of each transition or persistence divided by the total number of trees.
3 RESULTS
From the 144 SB0120-CdO strains selected in CdO and from the 219 SB0120-NAq strains selected in NAq, 123 and 290 SNPs were identified relative to the Mb3601 reference strain, respectively. When multiple isolates were available for a given outbreak, two or three unique SNPs sequences were detected in 39.7% of CdO outbreaks and 51.7% of NAq outbreaks.
3.1 Bayesian phylogenetic estimates
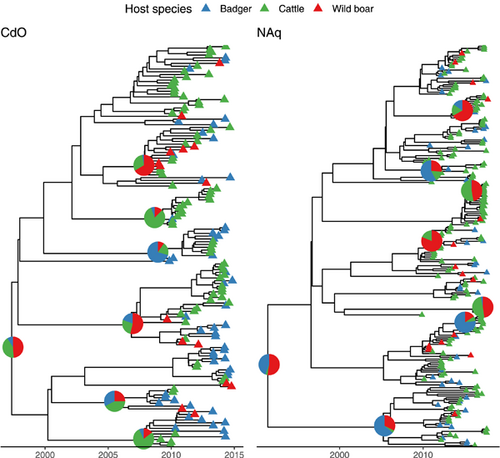
According to the parameters, trace inspection showing stationary distribution, and ESS < 200, all models fitted the data well. The best-fitted model, was an HKY substitution model, with an uncorrelated lognormal molecular clock for both study areas (Table A2). The corresponding inferred maximum clade credibility trees are shown in Figure 2.
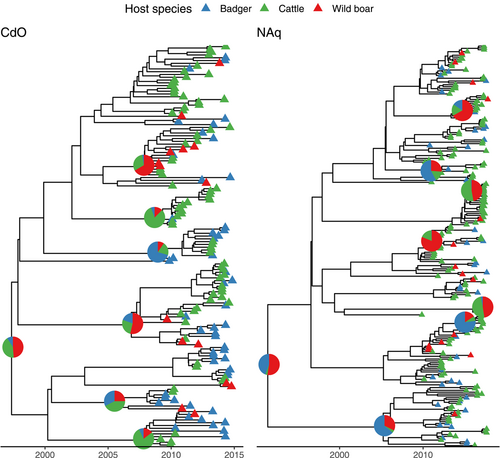
According to the best-fitting model for CdO, the most recent common ancestor (MRCA) of the studied isolates circulated in 1997 (with a 95% credibility interval between 1989 and 2002), as suggested by the median tree height (17.5 years, 95% highest posterior density (HPD): [12.6, 25.1]). It is however unclear whether the MRCA was hosted by wildlife or cattle (median host probability: 0.49 (95% HPD = [0, 0.97]) for wild boar, 0.35 (95% HPD = [0, 0.97]) for cattle, and 0.05 (95% HPD = [0, 0.33]) for badger). In NAq, the best-fitting model predicted that the MRCA of the studied isolates circulated in 1991 (with a 95% credibility interval between 1975 and 1999), according to the median tree height (25.4 years, 95% HPD: [18.1, 42.0]). While the HPD intervals are overlapping, the predicted MRCA would have been hosted in wildlife (host probability: 0.55 (95% HPD = [0.02; 0.94]) for wild boar, vs. 0.40 (95% HPD = [0.01; 0.92]) for badger and 0.02 (95% HPD = [0; 0.11]) for cattle). The mean molecular clock was significantly (p < 0.001, Wilcoxon test) smaller in CdO with a median estimate of 0.42 substitution/genome/year (95%HPD: [0.31, 0.54]) versus 0.57 substitution/genome/year (95% HPD: [0.44, 0.71]) for NAq.
For both study areas, we inferred that the effective population size was the largest for badgers (CdO: median of 9.4, 95% HPD: [4.9, 14.6]; NAq: median of 21.2, 95% HPD: [13.4, 29.6]) before cattle (CdO: median of 3.8, 95% HPD: [1.6, 6.6]; NAq: median of 5.0, 95%HPD: [1.8, 9.6]) and wild boars (CdO: median of 2.7, 95% HPD: [0.3, 7.0]; Naq: median of 2.0, 95% HPD: [0.3, 5.3]). This means that two lineages hosted by badgers are expected to coalesce more slowly than two lineages hosted by cattle or wild boar because the coalescent rate is the inverse of the effective population size.
3.2 Host species transition
Table 1 shows the proportion of transition rates selected (i.e., different from zero) by the BSSVS procedure as well as the inferred transition rates median and HPDs when these rates were selected. We defined the selection threshold as the proportion of transition rates selected by the BSSVS procedure. For example, a selection threshold of 0.5 for a given transition rate means that this transition rate was selected by the BSSVS procedure in half of the outputs. Considering an arbitrary selection threshold of 0.80, three transition rates were selected in CdO, namely from cattle to badgers, from badgers to wild boars, and from wild boars to cattle, and four in NAq which were the same as in CdO with the additional transition rate from badgers to cattle.
| Transition rate | CdO | NAq | ||||
|---|---|---|---|---|---|---|
| Selected | Median | HPD | Selected | Median | HPD | |
| From cattle to badgers | 0.99 | 0.27 | 0.08–0.54 | 0.80 | 0.17 | 0.03–0.36 |
| From wild boars to badgers | 0.58 | 0.09 | 0.01–0.47 | 0.41 | 0.11 | 0.01–0.30 |
| From badgers to cattle | 0.63 | 0.09 | 0.01–0.40 | 0.88 | 0.56 | 0.11–1.32 |
| From wild boars to cattle | 0.94 | 0.24 | 0.03–0.84 | 0.99 | 1.03 | 0.31–2.57 |
| From badgers to wild boars | 0.97 | 0.24 | 0.03–0.83 | 0.90 | 0.42 | 0.08–1.16 |
| From cattle to wild boars | 0.60 | 0.13 | 0.01–0.62 | 0.42 | 0.25 | 0.01–1.07 |
- Note: “Selected” represents the proportion transition rate selected by the BSSVS procedure (i.e., non-zero). The median and HPD were computed for the selected transition rates.
- Abbreviation: HPD, 95% highest probability distribution.
In both study areas, the inferred transition rates showed large variations. It is therefore difficult to conclude the frequency of each transition event. Globally, these results suggest that M. bovis migrated from cattle to badgers, from badgers to wild boars, and from wild boars to cattle. In addition, in NAq a transition back from badgers to cattle was also predicted.
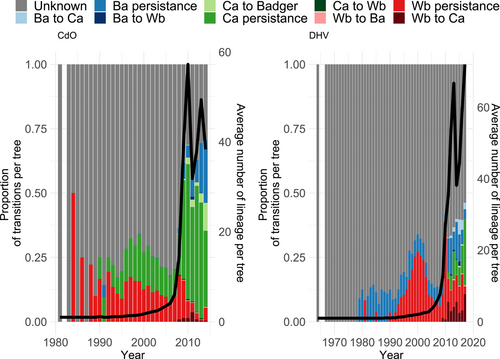
The proportion of unknown events, when the host species is considered unknown for a probability <0.9, is more important in NAq (0.89) than in CdO (0.36). For the known events, the majority is species persistence for both study areas with 0.90 in CdO (with 89.2% of persistence events being cattle persistence, 8.4% being badger persistence and 2.4% being wild boar persistence) and 0.88 in NAq (with 50.7% of persistence events being cattle persistence, 19.3% being badger persistence and 30.0% being wild boar persistence). Consistently with the inferred transition rate, the most frequent between-species transition was cattle-to-badger which represented 95.2% of all between-species transition events in CdO, and wild boar-to-cattle which represented 83.4% of all between-species transition events in NAq. These results suggest that between-species transitions remain rare events. Figure 3 shows the evolution with time of the average number of lineages and the proportion of transitions and persistence events per tree for both study areas. Moreover, it is worth mentioning that persistence events changed over time. Indeed, for both study areas, cattle persistence was most frequent and while badger persistence is identified onward starting as early as 1979 in NAq, it is gaining importance starting in 2008 only in CdO. On the opposite, cattle persistence is identified first in 1990 in CdO while only in 2009 in NAq.
4 DISCUSSION
In this study, we studied in two distinct study areas the transition of two different M. bovis strains belonging to the Eu3 clonal group among three host species, namely cattle, badgers, and wild boars. For this purpose, we used a structured coalescent model to infer transition rates between these three subpopulations. We showed that while the transition events remain rare events, our model predictions suggest that wild boars may be intermediaries for the transmission of M. bovis from badgers to cattle in both CdO and NAq.
We selected the same Bayesian evolutionary models for both study areas. More specifically, we selected an HKY substitution model and a lognormal molecular clock. The HKY substitution model has been previously used to model M. bovis phylogeny in cattle and brushtail possums in New Zealand (Crispell et al., 2017) and elks, white-tailed deer, and cattle in Michigan, USA (Salvador et al., 2019). We estimated two significantly different substitution rates for both regions. Both estimates were higher than previously estimated in Northern Ireland from cattle and badgers (Biek et al., 2012; Trewby et al., 2016) and in Michigan from cattle and elk (Salvador et al., 2019). However, the substitution rates estimated from cattle and possums in New Zealand are in the same order of magnitude as the one we estimated from NAq (Crispell et al., 2017) and the one estimated from the South-West of France from cattle and badgers (Duault et al., 2022) is in the same order of magnitude as the one we estimated for CdO. While, as noted by Duault et al., the differences between the studies could result from the different M. bovis lineage or the sampled species (Duault et al., 2022), this does not explain the significantly higher substitution rates inferred in NAq than in CdO. Indeed, for both study areas, the same host species were sampled during the same period. However, even if the spoligotype was the same in both study areas, the VNTR profiles differed. Hauer et al. showed that while SB0120 spoligotype was found all over France, specific VNTR profiles that spread locally were identified (such as 5 5 4 3 11 4 5 6 in CdO and 5 3 5 3 9 4 5 6 in NAq) (Hauer et al., 2015). While belonging to the Eu3 clonal group and sharing the same spoligotype, these strains were identified on different phylogenetic branches (Hauer et al., 2019) suggesting that VNTR profile and substitution rates could represent specific lineage characteristics.
To infer the internal nodes for host species, we used a structured coalescent method, which limits the impact of sampling bias (Müller et al., 2017). Indeed, contrary to the migration method (Lemey et al., 2009), structured coalescent methods do not assume that the migration process and tree-generating process are independent. According to the selected transition rates between badgers and cattle, the relationship between both species was clearly different between the two study areas: transition rate from cattle to badgers was more frequently predicted than from badgers to cattle in CdO, contrary to NAq where both transition rates were predicted in more than 80% of the outputs. This could relate to the implementation of bTB biosecurity measures toward wildlife that were more stringent in CdO than in NAq. Recent studies including samples collected from two species, namely badger and cattle, are in favor of the transition from badger to cattle (Crispell et al., 2019; Duault et al., 2022; van Tonder et al., 2021), however in one of these studies, while the overall badger-to-cattle transition rate was higher than the cattle-to-badger transition rate, a more refined analysis on transmission cluster levels revealed that for 4/12 clusters, cattle-to-badger transition rates were higher than badger-to-cattle transition ones (van Tonder et al., 2021). Moreover, an epidemiological study reconstructing the contact network between badger setts and cattle farms concluded the intermediary role of badgers in M. bovis transmission in the South-West of France (Bouchez-Zacria et al., 2018). In addition, according to the French graphic parcel register (Relevé parcellaire Graphique, https://www.geoportail.gouv.fr), the landscape in NAq is more fragmented with numerous interfaces between pastures and wooded areas, than in CdO. Fragmented landscapes were shown to be associated with lower adult badger population densities, which was corroborated by their lower densities in NAq than in CdO (Jacquier et al., 2021). However, the increased interfaces between pastures and wooded areas could increase the contact rates between badgers and cattle in NAq (Bouchez-Zacria et al., 2017) and explain the transition back and forth between these two species in this area compared to CdO. Also, bTB apparent prevalence in badgers might have increased between 2014 and 2016–2017 in NAq, while it has decreased in CdO during the same time interval (Réveillaud et al., 2018). The lack of evidence for the badger-to-cattle transition could also result from a lower statistical power due to the different sampling schemes in CdO and NAq.
It is noteworthy that we studied for the first time a multi-host system including wild boars in addition to badgers and cattle. We showed that among the selected transition rate in both NAq and CdO, the transition rate from wild boar to cattle was identified, suggesting a relatively frequent transition from wild boar to cattle and the intermediary role played by wild boars between badgers and cattle. This highlight the important role played by wild boars in the spread of M. bovis to other species. This could be related to the large distances traveled by wild boars (Podgórski et al., 2013) compared to badgers (Byrne et al., 2014). In addition, cattle movement, even if less frequent, could also enhance M. bovis spread between distant farms (Palisson et al., 2016). Furthermore, in both study areas host species persistence was identified as the main event while between-species transition represented less than 12% of known events, highlighting that between-species transition remains a rare event.
To summarize, we showed that while wild boars played the role of intermediary host between badgers and cattle, the role of badgers differed between both regions: in CdO badgers were intermediaries from cattle to wild boars, whereas in NAq, badgers transmitted to both cattle and wild boars. Several factors could explain these differences. Some of them are inherent to our study design, such as the lineage that can lead to different substitution rates, the temporal depth, or the number of strains collected, while other relates to the landscape or the population density of wildlife species.
Our work has several limitations. First, we excluded red foxes, red deer, and roe deer samples from our study because of the limited number of available samples, which prevented us from evaluating their role in this multi-host system. BTB-infected foxes (Vulpes vulpes) were detected in Dordogne in 2015 (Michelet et al., 2018). A further study showed that in Dordogne, Landes, and Charente, bTB prevalence in foxes ranged between 5% and 10%, similar to that observed in badgers and wild boars (Richomme et al., 2020). However, due to the lack of information concerning red foxes, we cannot conclude about their role. Second, the sampling procedure varies between cattle and wildlife, with nearly exhaustive testing of cattle while samples collected from wildlife depend on events such as hunting or road kills leading to an underestimation of M. bovis infection prevalence in wildlife. In addition, wildlife carcasses are subject to contamination and deterioration lowering the culture sensitivity and thus poorer statistical representativeness in our sample compared to cattle (Rivière et al., 2015). While inference with structured coalescent models is less altered by sampling bias, unsampled demes (such as the red foxes or cervids in our study) could reshape our results. Thirdly, our approach did not allow the inclusion of spatialized information that could describe the localization of sampled isolates or the different sizes of the home range for badgers and wild boars, or the change of pastures for cattle. Finally, a large number of transition events was labeled as unknown. This is particularly true for NAq where nearly 90% of transition could not be labeled. The results concerning host species persistence and between-species transition should therefore be considered with caution.
5 CONCLUSIONS
In conclusion, using a Bayesian evolutionary model, we inferred transition rates between cattle, badgers, and wild boars. Although this approach does not allow us to quantify within-species transmission, our result shed light on the wild boar role, which appears to act as an intermediary between badgers and cattle in the circulation of two different Eu3 M. bovis in two distinct study areas.
AUTHOR CONTRIBUTIONS
Laetitia Canini: Data curation (equal); formal analysis (equal); methodology (equal); validation (equal); visualization (equal); writing – original draft (equal). Gabriela Modenesi: Data curation (supporting); formal analysis (supporting); visualization (supporting); writing – original draft (supporting). Aurélie Courcoul: Conceptualization (equal); funding acquisition (equal); writing – original draft (equal). Maria-Laura Boschiroli: Conceptualization (equal); project administration (equal); resources (equal); supervision (equal); writing – original draft (equal). Benoit Durand: Conceptualization (equal); funding acquisition (equal); supervision (equal); writing – original draft (equal). Lorraine Michelet: Conceptualization (equal); formal analysis (equal); funding acquisition (equal); investigation (equal); methodology (equal); project administration (equal); writing – original draft (equal).
ACKNOWLEDGMENTS
The French Ministry of Agriculture financed the sampling and the analyses in the framework of the RFSA call on TB projects (Anses‑DGAl credit agreement no 170131).
CONFLICT OF INTEREST
None declared.
ETHICS STATEMENT
None required.
APPENDIX
| DA | Sample | Collection date | Host scientific name | Biosample | Accession |
|---|---|---|---|---|---|
| 10-00086-00 | 10Z000099 | 2009-11-09 | Bos taurus | SAMEA8955321 | ERR6198117 |
| 10-00566-00 | 10Z002121 | 2010-02-12 | Bos taurus | SAMEA8955322 | ERR6198204 |
| 10-01026-00 | 10Z005043 | 2010-03-26 | Bos taurus | SAMEA8955323 | ERR6198207 |
| D-10-02190 | 10Z008544 | 2010-09-18 | Sus scrofa | SAMEA8955324 | ERR6198208 |
| D-10-02191 | 10Z008545 | 2010-09-10 | Bos taurus | SAMEA8955325 | ERR6198209 |
| D-10-02192 | 10Z008546 | 2010-08-15 | Sus scrofa | SAMEA8955326 | ERR6198210 |
| D-10-02315 | 10Z009059 | 2010-09-12 | Sus scrofa | SAMEA8955327 | ERR6198216 |
| D-10-02451 | 10Z009733 | 2010-10-18 | Meles meles | SAMEA8955328 | ERR6198228 |
| D-11-00916 | 11Z001983 | 2011-01-04 | Bos taurus | SAMEA8955329 | ERR6198247 |
| D-11-01150 | 11Z002149 | 2011-02-08 | Bos taurus | SAMEA8955330 | ERR6198291 |
| D-11-01158 | 11Z002157 | 2011-02-27 | Meles meles | SAMEA8955331 | ERR6198317 |
| D-11-01374 | 11Z002528 | 2011-02-22 | Bos taurus | SAMEA8955332 | ERR6198349 |
| D-11-01375 | 11Z002529 | 2011-03-01 | Bos taurus | SAMEA8955333 | ERR6198365 |
| D-11-01905 | 11Z003570 | 2011-04-12 | Bos taurus | SAMEA8955334 | ERR6198374 |
| D-11-01992 | 11Z003734 | 2011-05-03 | Bos taurus | SAMEA8955335 | ERR6198375 |
| D-11-02082 | 11Z003894 | 2011-05-05 | Bos taurus | SAMEA8955336 | ERR6198376 |
| D-11-03278 | 11Z006170 | 2011-10-10 | Bos taurus | SAMEA8955337 | ERR6198377 |
| D-12-00020 | 12Z000029 | 2011-10-10 | Meles meles | SAMEA8955338 | ERR6198378 |
| D-12-01240 | 12Z002503 | 2012-01-11 | Meles meles | SAMEA8955339 | ERR6198379 |
| D-12-01581 | 12Z003372 | 2012-01-17 | Bos taurus | SAMEA8955340 | ERR6198389 |
| D-12-01582 | 12Z003373 | 2013-01-22 | Meles meles | SAMEA8955341 | ERR6198390 |
| D-12-01589 | 12Z003430 | 2012-03-12 | Sus scrofa | SAMEA8955342 | ERR6198391 |
| D-12-01597 | 12Z003438 | 2012-01-27 | Bos taurus | SAMEA8955343 | ERR6198393 |
| D-12-01601 | 12Z003442 | 2012-01-24 | Bos taurus | SAMEA8955344 | ERR6198394 |
| D-12-01697 | 12Z003519 | 2012-02-14 | Bos taurus | SAMEA8955345 | ERR6198395 |
| D-12-01854 | 12Z004182 | 2012-02-13 | Meles meles | SAMEA8955346 | ERR6198396 |
| D-12-01929 | 12Z004424 | 2012-02-14 | Bos taurus | SAMEA8955347 | ERR6198398 |
| D-12-02001 | 12Z004494 | 2012-03-16 | Bos taurus | SAMEA8955348 | ERR6198399 |
| D-12-02007 | 12Z004505 | 2012-02-09 | Bos taurus | SAMEA8955349 | ERR6198400 |
| D-12-02120 | 12Z004741 | 2012-03-23 | Bos taurus | SAMEA8955350 | ERR6198401 |
| D-12-02121 | 12Z004742 | 2012-02-28 | Bos taurus | SAMEA8955351 | ERR6198442 |
| D-12-02129 | 12Z004749 | 2012-03-13 | Bos taurus | SAMEA8955352 | ERR6198443 |
| D-12-02131 | 12Z004751 | 2012-03-13 | Bos taurus | SAMEA8955353 | ERR6198444 |
| D-12-02240 | 12Z004928 | 2012-03-16 | Bos taurus | SAMEA8955354 | ERR6198445 |
| D-12-02242 | 12Z004930 | 2012-03-27 | Bos taurus | SAMEA8955355 | ERR6198447 |
| D-12-02292 | 12Z004995 | 2012-03-07 | Meles meles | SAMEA8955356 | ERR6198448 |
| D-12-02293 | 12Z004996 | 2012-03-22 | Bos taurus | SAMEA8955357 | ERR6198449 |
| D-12-02296 | 12Z004999 | 2012-04-02 | Meles meles | SAMEA8955358 | ERR6199027 |
| D-12-02372 | 12Z005163 | 2012-04-06 | Meles meles | SAMEA8955359 | ERR6201794 |
| D-12-02472 | 12Z005339 | 2012-04-05 | Meles meles | SAMEA8955360 | ERR6201795 |
| D-12-02749 | 12Z005660 | 2012-05-05 | Meles meles | SAMEA8955361 | ERR6201796 |
| D-12-02971 | 12Z006182 | 2012-05-22 | Bos taurus | SAMEA8955362 | ERR6201797 |
| D-12-03180 | 12Z006994 | 2012-05-09 | Bos taurus | SAMEA8955363 | ERR6201798 |
| D-12-03307 | 12Z007111 | 2013-05-24 | Meles meles | SAMEA8955364 | ERR6201800 |
| D-12-03422 | 12Z007219 | 2012-07-23 | Bos taurus | SAMEA8955365 | ERR6201801 |
| D-12-03423 | 12Z007220 | 2012-07-20 | Bos taurus | SAMEA8955366 | ERR6201802 |
| D-13-00134 | 13Z000269 | 2012-11-16 | Meles meles | SAMEA8955367 | ERR6201804 |
| D-13-00505 | 13Z001275 | 2013-01-03 | Meles meles | SAMEA8955368 | ERR6201806 |
| D-13-00830 | 13Z001729 | 2013-01-25 | Bos taurus | SAMEA8955369 | ERR6201807 |
| D-13-00831 | 13Z001730 | 2013-01-25 | Bos taurus | SAMEA8955370 | ERR6201808 |
| D-13-00956 | 13Z001898 | 2013-02-01 | Bos taurus | SAMEA8955371 | ERR6201809 |
| D-13-00962 | 13Z001903 | 2013-02-15 | Bos taurus | SAMEA8955372 | ERR6201810 |
| D-13-01061 | 13Z001990 | 2013-01-02 | Bos taurus | SAMEA8955373 | ERR6201811 |
| D-13-01238 | 13Z002208 | 2013-02-20 | Bos taurus | SAMEA8955374 | ERR6201812 |
| D-13-01245 | 13Z002215 | 2013-02-15 | Bos taurus | SAMEA8955375 | ERR6201813 |
| D-13-01303 | 13Z002277 | 2013-03-08 | Bos taurus | SAMEA8955376 | ERR6201814 |
| D-13-01556 | 13Z002699 | 2013-03-29 | Bos taurus | SAMEA8955377 | ERR6201815 |
| D-13-01562 | 13Z002705 | 2013-04-12 | Bos taurus | SAMEA8955378 | ERR6201817 |
| D-13-01565 | 13Z002708 | 2013-04-22 | Bos taurus | SAMEA8955379 | ERR6201818 |
| D-13-01566 | 13Z002709 | 2013-04-22 | Bos taurus | SAMEA8955380 | ERR6201819 |
| D-13-02048 | 13Z003478 | 2013-06-05 | Bos taurus | SAMEA8955381 | ERR6201820 |
| D-13-02100 | 13Z003643 | 2013-04-23 | Bos taurus | SAMEA8955382 | ERR6201821 |
| D-13-02101 | 13Z003644 | 2013-06-07 | Bos taurus | SAMEA8955383 | ERR6201822 |
| D-13-02225 | 13Z003769 | 2013-03-22 | Bos taurus | SAMEA8955384 | ERR6201823 |
| D-13-02226 | 13Z003770 | 2013-06-13 | Bos taurus | SAMEA8955385 | ERR6201825 |
| D-13-02368 | 13Z004012 | 2013-06-13 | Bos taurus | SAMEA8955386 | ERR6201826 |
| D-13-02401 | 13Z004051 | 2013-06-13 | Bos taurus | SAMEA8955387 | ERR6201828 |
| D-13-02436 | 13Z004129 | 2013-04-23 | Bos taurus | SAMEA8955388 | ERR6201829 |
| D-13-02620 | 13Z004667 | 2013-06-13 | Bos taurus | SAMEA8955389 | ERR6201830 |
| D-13-02704 | 13Z004785 | 2013-08-06 | Bos taurus | SAMEA8955390 | ERR6201831 |
| D-13-03134 | 13Z005631 | 2013-06-13 | Bos taurus | SAMEA8955391 | ERR6201832 |
| D-13-03137 | 13Z005634 | 2013-08-22 | Sus scrofa | SAMEA8955392 | ERR6201833 |
| D-13-03140 | 13Z005637 | 2013-09-15 | Sus scrofa | SAMEA8955393 | ERR6201835 |
| D-13-03227 | 13Z005787 | 2013-09-03 | Bos taurus | SAMEA8955394 | ERR6201836 |
| D-13-03897 | 13Z010139 | 2013-10-29 | Bos taurus | SAMEA8955395 | ERR6201837 |
| D-13-03898 | 13Z010140 | 2013-11-15 | Bos taurus | SAMEA8955396 | ERR6201838 |
| D-13-03899 | 13Z010141 | 2013-10-13 | Sus scrofa | SAMEA8955397 | ERR6201839 |
| D-13-03900 | 13Z010142 | 2013-11-10 | Sus scrofa | SAMEA8955398 | ERR6201840 |
| D-14-00007 | 14Z000006 | 2013-11-03 | Sus scrofa | SAMEA8955399 | ERR6201841 |
| D-14-00013 | 14Z000010 | 2013-11-20 | Bos taurus | SAMEA8955400 | ERR6201842 |
| D-14-00016 | 14Z000013 | 2013-11-19 | Bos taurus | SAMEA8955401 | ERR6201843 |
| D-14-00019 | 14Z000015 | 2013-11-19 | Bos taurus | SAMEA8955402 | ERR6201844 |
| D-14-00020 | 14Z000016 | 2013-11-19 | Bos taurus | SAMEA8955403 | ERR6201845 |
| D-14-01072 | 14Z003302 | 2013-12-22 | Meles meles | SAMEA8955404 | ERR6201847 |
| D-14-01074 | 14Z003304 | 2014-01-13 | Meles meles | SAMEA8955405 | ERR6201848 |
| D-14-01623 | 14Z004188 | 2014-03-28 | Bos taurus | SAMEA8955406 | ERR6201849 |
| D-14-01757 | 14Z004539 | 2014-03-13 | Bos taurus | SAMEA8955407 | ERR6201850 |
| D-14-01758 | 14Z004540 | 2014-03-12 | Bos taurus | SAMEA8955408 | ERR6201851 |
| D-14-02086 | 14Z005338 | 2014-04-28 | Bos taurus | SAMEA8955409 | ERR6201852 |
| D-14-02099 | 14Z005351 | 2014-04-23 | Bos taurus | SAMEA8955410 | ERR6201853 |
| D-14-02112 | 14Z005364 | 2014-04-23 | Bos taurus | SAMEA8955411 | ERR6201854 |
| D-14-02116 | 14Z005365 | 2014-04-09 | Bos taurus | SAMEA8955412 | ERR6201855 |
| D-14-02259 | 14Z005576 | 2014-05-06 | Meles meles | SAMEA8955413 | ERR6201857 |
| D-14-02574 | 14Z006089 | 2014-05-25 | Meles meles | SAMEA8955414 | ERR6201858 |
| D-14-02811 | 14Z006436 | 2014-06-23 | Bos taurus | SAMEA8955415 | ERR6201860 |
| D-14-02871 | 14Z006581 | 2014-07-24 | Bos taurus | SAMEA8955416 | ERR6201861 |
| D-14-03006 | 14Z006798 | 2014-06-05 | Meles meles | SAMEA8955417 | ERR6201862 |
| D-14-03118 | 14Z007047 | 2014-06-05 | Meles meles | SAMEA8955419 | ERR6201863 |
| D-14-03380 | 14Z007947 | 2014-09-10 | Bos taurus | SAMEA8955420 | ERR6201864 |
| D-14-03463 | 14Z008062 | 2014-09-10 | Bos taurus | SAMEA8955421 | ERR6201865 |
| D-14-03644 | 14Z008376 | 2014-09-17 | Bos taurus | SAMEA8955422 | ERR6201866 |
| D-14-03697 | 14Z008498 | 2014-08-24 | Sus scrofa | SAMEA8955423 | ERR6201868 |
| D-14-04057 | 14Z009345 | 2014-09-21 | Sus scrofa | SAMEA8955424 | ERR6201869 |
| D-15-00525 | 15Z001806 | 2015-01-14 | Bos taurus | SAMEA8955425 | ERR6201870 |
| D-15-00613 | 15Z002145 | 2015-01-23 | Bos taurus | SAMEA8955426 | ERR6201871 |
| D-15-00828 | 15Z002493 | 2015-02-13 | Bos taurus | SAMEA8955427 | ERR6201872 |
| D-15-00970 | 15Z002980 | 2015-02-17 | Bos taurus | SAMEA8955428 | ERR6201873 |
| D-15-01174 | 15Z004066 | 2015-03-06 | Bos taurus | SAMEA8955429 | ERR6201874 |
| D-15-01176 | 15Z004068 | 2015-03-20 | Bos taurus | SAMEA8955430 | ERR6201875 |
| D-15-01301 | 15Z004334 | 2015-01-25 | Meles meles | SAMEA8955431 | ERR6201877 |
| D-15-01302 | 15Z004335 | 2015-02-27 | Bos taurus | SAMEA8955432 | ERR6201878 |
| D-15-01311 | 15Z004344 | 2015-03-30 | Meles meles | SAMEA8955433 | ERR6201879 |
| D-15-01312 | 15Z004345 | 2015-04-15 | Bos taurus | SAMEA8955434 | ERR6201881 |
| D-15-01346 | 15Z004377 | 2015-03-12 | Meles meles | SAMEA8955435 | ERR6201882 |
| D-15-01460 | 15Z004939 | 2015-03-13 | Meles meles | SAMEA8955436 | ERR6201883 |
| D-15-01557 | 15Z005359 | 2015-03-22 | Meles meles | SAMEA8955437 | ERR6201884 |
| D-15-01665 | 15Z005833 | 2015-04-17 | Bos taurus | SAMEA8955438 | ERR6201885 |
| D-15-01667 | 15Z005835 | 2013-05-21 | Meles meles | SAMEA8955439 | ERR6201886 |
| D-15-01759 | 15Z006243 | 2015-05-18 | Bos taurus | SAMEA8955440 | ERR6201887 |
| D-15-01919 | 15Z006492 | 2015-04-08 | Meles meles | SAMEA8955441 | ERR6201888 |
| D-15-01922 | 15Z006495 | 2015-05-18 | Bos taurus | SAMEA8955442 | ERR6201889 |
| D-15-01960 | 15Z006541 | 2015-05-12 | Meles meles | SAMEA8955443 | ERR6201890 |
| D-15-02070 | 15Z006760 | 2015-06-17 | Meles meles | SAMEA8955444 | ERR6201891 |
| D-15-03139 | 15Z010210 | 2015-10-14 | Bos taurus | SAMEA8955445 | ERR6201893 |
| D-15-03409 | 15Z010962 | 2015-09-13 | Sus scrofa | SAMEA8955446 | ERR6201894 |
| D-16-00172 | 16Z000595 | 2015-12-15 | Bos taurus | SAMEA8955447 | ERR6201895 |
| D-16-00301 | 16Z000964 | 2015-12-13 | Sus scrofa | SAMEA8955448 | ERR6201896 |
| D-16-00302 | 16Z000965 | 2015-12-26 | Sus scrofa | SAMEA8955449 | ERR6201897 |
| D-16-00361 | 16Z001097 | 2015-12-15 | Bos taurus | SAMEA8955450 | ERR6201898 |
| D-16-01021 | 16Z002887 | 2016-02-15 | Bos taurus | SAMEA8955451 | ERR6201899 |
| D-16-01467 | 16Z004107 | 2016-03-11 | Bos taurus | SAMEA8955452 | ERR6201901 |
| D-16-01566 | 16Z004271 | 2016-03-25 | Bos taurus | SAMEA8955453 | ERR6201902 |
| D-16-01567 | 16Z004272 | 2016-03-20 | Meles meles | SAMEA8955454 | ERR6201903 |
| D-16-01569 | 16Z004274 | 2016-03-10 | Bos taurus | SAMEA8955455 | ERR6201906 |
| D-16-01664 | 16Z004560 | 2016-03-22 | Bos taurus | SAMEA8955456 | ERR6201907 |
| D-16-01669 | 16Z004566 | 2016-03-08 | Bos taurus | SAMEA8955457 | ERR6201908 |
| D-16-01672 | 16Z004569 | 2016-02-17 | Meles meles | SAMEA8955458 | ERR6201909 |
| D-16-01723 | 16Z004699 | 2016-03-08 | Meles meles | SAMEA8955459 | ERR6201910 |
| D-16-01847 | 16Z005204 | 2016-04-12 | Bos taurus | SAMEA8955460 | ERR6201915 |
| D-16-01848 | 16Z005206 | 2016-04-05 | Bos taurus | SAMEA8955461 | ERR6201937 |
| D-16-01849 | 16Z005207 | 2016-04-06 | Bos taurus | SAMEA8955462 | ERR6201945 |
| D-16-01851 | 16Z005209 | 2016-03-25 | Meles meles | SAMEA8955463 | ERR6201960 |
| D-16-01852 | 16Z005210 | 2016-04-11 | Bos taurus | SAMEA8955464 | ERR6201972 |
| D-16-02216 | 16Z006257 | 2017-03-27 | Meles meles | SAMEA8955465 | ERR6201978 |
| D-16-02405 | 16Z006707 | 2016-05-27 | Bos taurus | SAMEA8955466 | ERR6201986 |
| D-16-02601 | 16Z007121 | 2016-05-10 | Meles meles | SAMEA8955467 | ERR6201998 |
| D-16-02833 | 16Z007476 | 2016-06-20 | Bos taurus | SAMEA8955468 | ERR6202004 |
| D-16-02834 | 16Z007477 | 2016-06-24 | Bos taurus | SAMEA8955469 | ERR6202040 |
| D-16-02835 | 16Z007478 | 2016-06-27 | Bos taurus | SAMEA8955470 | ERR6202401 |
| D-16-02836 | 16Z007479 | 2016-07-01 | Bos taurus | SAMEA8955471 | ERR6203392 |
| D-16-02924 | 16Z007681 | 2016-06-20 | Bos taurus | SAMEA8955472 | ERR6204677 |
| D-16-03225 | 16Z008197 | 2016-07-21 | Bos taurus | SAMEA8955473 | ERR6206178 |
| D-16-03320 | 16Z008443 | 2016-07-17 | Meles meles | SAMEA8955474 | ERR6208726 |
| D-17-00049 | 17Z000058 | 2016-12-01 | Bos taurus | SAMEA8955475 | ERR6208976 |
| D-17-00050 | 17Z000059 | 2016-12-01 | Bos taurus | SAMEA8955476 | ERR6209044 |
| D-17-00051 | 17Z000060 | 2016-12-01 | Bos taurus | SAMEA8955477 | ERR6209102 |
| D-17-00055 | 17Z000064 | 2016-11-01 | Sus scrofa | SAMEA8955478 | ERR6209192 |
| D-17-00269 | 17Z000349 | 2016-12-06 | Bos taurus | SAMEA8955479 | ERR6209278 |
| D-17-00270 | 17Z000350 | 2016-12-06 | Bos taurus | SAMEA8955480 | ERR6209301 |
| D-17-00270 | 17Z000351 | 2016-12-06 | Bos taurus | SAMEA8955481 | ERR6209309 |
| D-17-00540 | 17Z000898 | 2016-12-01 | Bos taurus | SAMEA8955482 | ERR6209323 |
| D-17-00563 | 17Z000914 | 2017-01-09 | Bos taurus | SAMEA8955483 | ERR6209335 |
| D-17-00641 | 17Z001046 | 2017-01-20 | Bos taurus | SAMEA8955484 | ERR6209343 |
| D-17-00710 | 17Z001177 | 2017-01-09 | Bos taurus | SAMEA8955485 | ERR6209344 |
| D-17-00849 | 17Z001576 | 2017-02-03 | Bos taurus | SAMEA8955486 | ERR6209345 |
| D-17-00850 | 17Z001577 | 2017-02-03 | Bos taurus | SAMEA8955487 | ERR6209346 |
| D-17-00851 | 17Z001578 | 2017-02-02 | Bos taurus | SAMEA8955488 | ERR6209347 |
| D-17-00854 | 17Z001581 | 2017-02-02 | Bos taurus | SAMEA8955489 | ERR6209348 |
| D-17-01066 | 17Z002146 | 2017-02-07 | Bos taurus | SAMEA8955490 | ERR6209349 |
| D-17-01071 | 17Z002151 | 2017-02-10 | Bos taurus | SAMEA8955491 | ERR6209350 |
| D-17-01146 | 17Z002194 | 2017-02-15 | Bos taurus | SAMEA8955492 | ERR6209351 |
| D-17-01148 | 17Z002196 | 2017-02-09 | Bos taurus | SAMEA8955493 | ERR6209352 |
| D-17-01149 | 17Z002197 | 2017-02-10 | Bos taurus | SAMEA8955494 | ERR6209353 |
| D-17-01150 | 17Z002198 | 2017-02-07 | Bos taurus | SAMEA8955495 | ERR6209422 |
| D-17-01286 | 17Z002553 | 2017-01-18 | Bos taurus | SAMEA8955496 | ERR6209423 |
| D-17-01306 | 17Z002558 | 2017-02-27 | Bos taurus | SAMEA8955497 | ERR6209424 |
| D-17-01412 | 17Z002740 | 2017-02-07 | Bos taurus | SAMEA8955498 | ERR6209443 |
| D-17-01416 | 17Z002744 | 2017-03-08 | Bos taurus | SAMEA8955499 | ERR6209444 |
| D-17-01517 | 17Z002865 | 2017-03-23 | Bos taurus | SAMEA8955500 | ERR6209445 |
| D-17-01610 | 17Z003109 | 2017-03-01 | Bos taurus | SAMEA8955501 | ERR6209491 |
| D-17-01611 | 17Z003110 | 2017-02-10 | Bos taurus | SAMEA8955502 | ERR6209492 |
| D-17-01695 | 17Z003235 | 2017-04-05 | Bos taurus | SAMEA8955503 | ERR6209493 |
| D-17-01696 | 17Z003236 | 2017-03-28 | Bos taurus | SAMEA8955504 | ERR6209524 |
| D-17-01698 | 17Z003238 | 2017-03-21 | Bos taurus | SAMEA8955505 | ERR6209526 |
| D-17-01705 | 17Z003245 | 2017-03-27 | Bos taurus | SAMEA8955506 | ERR6209527 |
| D-17-01929 | 17Z003568 | 2017-03-30 | Bos taurus | SAMEA8955507 | ERR6209542 |
| D-17-01931 | 17Z003570 | 2017-04-13 | Bos taurus | SAMEA8955508 | ERR6209573 |
| D-17-02078 | 17Z004077 | 2017-04-21 | Bos taurus | SAMEA8955509 | ERR6209576 |
| D-17-02080 | 17Z004080 | 2017-04-20 | Bos taurus | SAMEA8955510 | ERR6210127 |
| D-17-02089 | 17Z004088 | 2017-04-20 | Bos taurus | SAMEA8955511 | ERR6210128 |
| D-17-02090 | 17Z004128 | 2017-04-19 | Bos taurus | SAMEA8955512 | ERR6210129 |
| D-17-02088 | 17Z004275 | 2017-04-20 | Bos taurus | SAMEA8955513 | ERR6210130 |
| D-17-02260 | 17Z004423 | 2017-03-29 | Bos taurus | SAMEA8955514 | ERR6210147 |
| D-17-02262 | 17Z004425 | 2017-04-19 | Bos taurus | SAMEA8955515 | ERR6210148 |
| D-17-02264 | 17Z004428 | 2017-05-02 | Bos taurus | SAMEA8955516 | ERR6210149 |
| D-17-02335 | 17Z004525 | 2017-04-24 | Bos taurus | SAMEA8955517 | ERR6210150 |
| D-17-02438 | 17Z005047 | 2017-04-26 | Bos taurus | SAMEA8955518 | ERR6210158 |
| D-17-02580 | 17Z005388 | 2017-04-19 | Bos taurus | SAMEA8955519 | ERR6210161 |
| D-17-02586 | 17Z005394 | 2017-05-09 | Bos taurus | SAMEA8955520 | ERR6210169 |
| D-17-02587 | 17Z005395 | 2017-05-18 | Bos taurus | SAMEA8955521 | ERR6210175 |
| D-17-02698 | 17Z005495 | 2017-05-22 | Bos taurus | SAMEA8955522 | ERR6210188 |
| D-17-02699 | 17Z005496 | 2017-05-22 | Bos taurus | SAMEA8955523 | ERR6210195 |
| D-17-02775 | 17Z005671 | 2017-05-30 | Bos taurus | SAMEA8955524 | ERR6210201 |
| D-17-02902 | 17Z005849 | 2017-06-23 | Bos taurus | SAMEA8955525 | ERR6210207 |
| D-17-03220 | 17Z006451 | 2017-03-13 | Meles meles | SAMEA8955526 | ERR6210212 |
| D-17-03217 | 17Z006454 | 2017-06-12 | Bos taurus | SAMEA8955527 | ERR6210216 |
| D-17-03251 | 17Z006498 | 2017-07-17 | Bos taurus | SAMEA8955528 | ERR6210223 |
| D-17-03430 | 17Z006770 | 2017-07-17 | Bos taurus | SAMEA8955529 | ERR6210230 |
| D-17-03431 | 17Z006771 | 2017-07-15 | Meles meles | SAMEA8955530 | ERR6210236 |
| D-17-03656 | 17Z007283 | 2017-08-06 | Bos taurus | SAMEA8955531 | ERR6210252 |
| D-17-03910 | 17Z007631 | 2017-08-24 | Bos taurus | SAMEA8955532 | ERR6210258 |
| D-17-03912 | 17Z007633 | 2017-08-10 | Bos taurus | SAMEA8955533 | ERR6210259 |
| D-17-03913 | 17Z007634 | 2017-08-10 | Bos taurus | SAMEA8955534 | ERR6210266 |
| D-17-04408 | 17Z008463 | 2017-08-24 | Bos taurus | SAMEA8955535 | ERR6210267 |
| D-17-04412 | 17Z008467 | 2017-07-09 | Meles meles | SAMEA8955536 | ERR6210268 |
| D-17-04421 | 17Z008476 | 2017-08-14 | Meles meles | SAMEA8955537 | ERR6210269 |
| D-17-04423 | 17Z008478 | 2017-04-27 | Meles meles | SAMEA8955538 | ERR6210270 |
| D-17-04936 | 17Z010330 | 2017-10-01 | Sus scrofa | SAMEA8955539 | ERR6210271 |
| D-17-05074 | 17Z010552 | 2017-10-29 | Sus scrofa | SAMEA8955540 | ERR6210272 |
| Model | Substitution model | Molecular clock model | Marginal likelihood | SD | BF | Reference model | |
|---|---|---|---|---|---|---|---|
| CdO | |||||||
| M1CdO | JC69 | Strict | −24554 | 0.09 | - | - | - |
| M2CdO | HKY | Strict | −24225 | 1.00 | 329 | 2.00 | M1CdO |
| M3CdO | GTR | Strict | Does not converge | ||||
| M4CdO | HKY | Uncorrelated lognormal | −24222 | 0.84 | 3 | 2.61 | M2CdO |
| M5CdO | HKY | Uncorrelated exponential | −24223 | 0.91 | −1 | 2.48 | M4CdO |
| NAq | |||||||
| M1NAq | JC69 | Strict | −87642 | 0.18 | - | - | - |
| M2NAq | HKY | Strict | −3400 | 9.41 | 84242 | 18.82 | M1NAq |
| M3NAq | GTR | Strict | Does not converge | ||||
| M4NAq | HKY | Uncorrelated lognormal | −3350 | 8.87 | 50 | 25.86 | M2NAq |
| M5NAq | HKY | Uncorrelated exponential | −87616 | 1.04 | −84266 | 17.9 | M4NAq |
- The marginal likelihood was computed with the Nested Sampling algorithm implemented in the BEAST NS package, with 10 particles. For all models, an unstructured coalescent population model was used. BF stands for Bayes Factor and was computed as ML1-ML2 if ML1 is the marginal likelihood of model 1 and ML2 marginal likelihood of model 2. To distinguish between both MLs, the difference should be greater than , where SD1 stands for the standard deviation of model 1 and SD2 standard deviation of model 2.
- Abbreviations: BF, Bayes factor; ML1, marginal likelihood of Model 1; ML2, marginal likelihood of Model 2; SD1, standard deviation of Model 1; SD2, SD2 standard deviation of Model 2.
Open Research
DATA AVAILABILITY STATEMENT
All data are provided in full in the results section of this paper apart from all WGS data which are available in NCBI GenBank under BioProject PRJEB46102 for NAq: https://www.ncbi.nlm.nih.gov/bioproject/PRJEB46102 and PRJEB46417 for CdO: https://www.ncbi.nlm.nih.gov/bioproject/PRJEB46417. The individual isolates can be accessed under the following BioSample accession numbers: SAMEA8955321 - SAMEA8955540 for NAq and SAMEA8987071 - SAMEA8987214 for CdO.