Comparison of immune-related gene signatures and immune infiltration features in early- and late-onset preeclampsia
Quanfeng Wu and Xiang Ying contributed equally to this work.
Abstract
Background
Preeclampsia, a severe pregnancy syndrome, is widely accepted divided into early- and late-onset preeclampsia (EOPE and LOPE) based on the onset time of preeclampsia, with distinct pathophysiological origins. However, the molecular mechanism especially immune-related mechanisms for EOPE and LOPE is currently obscure. In the present study, we focused on placental immune alterations between EOPE and LOPE and search for immune-related biomarkers that could potentially serve as potential therapeutic targets through bioinformatic analysis.
Methods
The gene expression profiling data was obtained from the Gene Expression Omnibus database. ESTIMATE algorithm and Gene Set Enrichment Analysis were employed to evaluate the immune status. The intersection of differentially expressed genes in GSE74341 series and immune-related genes set screened differentially expressed immune-related genes. Protein–protein interaction network and random forest were used to identify hub genes with a validation by a quantitative real-time PCR. Kyoto Encyclopedia of Genes and Genomes pathways, Gene Ontology and gene set variation analysis were utilized to conduct biological function and pathway enrichment analyses. Single-sample gene set enrichment analysis and CIBERSORTx tools were employed to calculate the immune cell infiltration score. Correlation analyses were evaluated by Pearson correlation analysis. Hub genes-miRNA network was performed by the NetworkAnalyst online tool.
Results
Immune score and stromal score were all lower in EOPE samples. The immune system-related gene set was significantly downregulated in EOPE compared to LOPE samples. Four hub differentially expressed immune-related genes (IL15, GZMB, IL1B and CXCL12) were identified based on a protein–protein interaction network and random forest. Quantitative real-time polymerase chain reaction validated the lower expression levels of four hub genes in EOPE compared to LOPE samples. Immune cell infiltration analysis found that innate and adaptive immune cells were apparent lacking in EOPE samples compared to LOPE samples. Cytokine-cytokine receptor, para-inflammation, major histocompatibility complex class I and T cell co-stimulation pathways were significantly deficient and highly correlated with hub genes. We constructed a hub genes-miRNA regulatory network, revealing the correlation between hub genes and hsa-miR-374a-5p, hsa-miR-203a-3p, hsa-miR-128-3p, hsa-miR-155-3p, hsa-miR-129-2-3p and hsa-miR-7-5p.
Conclusions
The innate and adaptive immune systems were severely impaired in placentas of EOPE. Four immune-related genes (IL15, GZMB, IL1B and CXCL12) were closely correlated with immune-related pathogenesis of EOPE. The result of our study may provide a new basis for discriminating between EOPE and LOPE and acknowledging the role of the immune landscape in the eventual interference and tailored treatment of EOPE.
1 INTRODUCTION
Preeclampsia is a severe pregnancy syndrome, which causing the deaths of > 70,000 pregnant woman and 500,00 perinatal fetus.1, 2 Preeclamptic women who survive are more likely to suffer from strokes, cardiovascular diseases and diabetes, which results in shorter lifespans. At the same time, perinatal fetus from preeclampsia pregnancy face a high risk of preterm birth, neurodevelopmental delay, and cardiovascular and metabolic disease in future life.3, 4 Although preeclampsia's symptoms have been documented for about 200 years, there is no effective treatment other than delivery. A major obstacle in the current treatment challenges for preeclampsia is how to detect placental dysfunction well before diagnosis because of difficulty in defining the molecular mechanism and potential therapeutic targets.2, 4 Early prediction and prevention are limited as a result of a poor understanding of the pathogenesis of such disorders. Therefore, further study into the pathogenesis of preeclampsia is critical to improve preeclampsia diagnosis and treatment.
Long-standing research has shown that preeclampsia eventually goes away if the placenta is removed, but not a fetus. It is therefore predominantly a placental issue in terms of pathogenesis.5 Although the pathogenesis is incompletely understood, preeclampsia is considered to have a two-stage pathophysiology. First, early in placental development, poor cytotrophoblast invasion and modification of spiral arterioles (pseudo-vasculogenesis), probably caused by a combination of genetic, immunologic and environmental factors, reduce uteroplacental blood flow. Then, systemic endothelial dysfunction is brought on by the ischemia and oxidative stress caused by the hypoxic placenta. As a result, the clinical features of preeclampsia spectrum disorder show multisystem end-organ damage.6 Preeclampsia is widely accepted to be divided into early-onset (onset time < 34 weeks of gestation) and late-onset (onset time ≥ 34 weeks of gestation) preeclampsia (EOPE and LOPE). Although LOPE may focus on interactions between placental oxidative stress and maternal genetic propensity to metabolic disorders and cardiovascular, EOPE is commonly considered to be a primary placental etiology.7 The difference of placental mRNA expression profiles between EOPE and LOPE further confirms the different origins of the two subtypes.8 Few studies have focused on the difference between EOPE and LOPE, which has lead to the contradictions of pathogenesis in preeclampsia.
Numerous studies in recent years have shown that the occurrence and progression of preeclampsia are strongly correlated with an immune system imbalance.9 Interactions between the mother and the placenta are significantly influenced by immune cells that accumulate within and around the organ in a healthy pregnancy during all trimesters. Innate immune system cells predominately occupy the decidua, the placental bed. These immune cells form a band around the trophoblasts and perform many functions. The immune system regulates trophoblast invasion, protects feto-maternal tolerance and combats encroaching infections if needed.10 Partial breakdown of immune tolerance results in the uterine spiral artery's inadequate metamorphosis, which causes a placenta abnormality. A healthy pregnancy depends on the balance of cytokines and immune cells produced at the maternal-fetal contact. An aberrant placental structure or angiogenesis may result from any local immune response imbalance.11 However, investigations on modifications of placental immune cells in preeclampsia have yielded conflicting results and the precise roles of immune cells in placenta have yet to be clarified. Immune maladaptation plays a very small role in LOPE, whereas it is widely assumed that the most common phenotype of preeclampsia is LOPE, and some researchers consider that preeclampsia is brought on by oxidative stress of the placenta, a poor metabolic or cardiovascular state prior to pregnancy. Scandinavian and US research that has focused exclusively on this type of preeclampsia challenged the immunological maladaptation hypothesis.12, 13 Although less common than LOPE, EOPE is associated with higher neonatal mortality and maternal morbidity.14 It is generally assumed that the pathogenesis of EOPE is the result of insufficient recasting of the uterine spiral artery, correlated with the immune system. Therefore, we consider that the immune system to be implicated in the pathogenesis of preeclampsia. However, the pathogenesis of EOPE is still far from being clarified and, especially, the molecular mechanism of immune maladaptation is still not well understood.
In the present study, we analyzed a preeclamspia datasets which contained EOPE, LOPE and normal sample from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo) database. Using gene set enrichment analysis (GSEA) and the ESTIMATE (i.e., Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data) algorithm, we revealed the impaired immune systems in EOPE, but not LOPE and normal samples. After acquiring differentially expressed immune-related genes between EOPE and LOPE samples, hub genes were identified using a protein–protein interaction (PPI) network and random forest (RF) and a quantitative real-time PCR (qRT-PCR) validated the lower expression levels of hub genes in EOPE compared to LOPE samples. Then, CIBERSORTx (https://cibersortx.stanford.edu) and single-sample GSEA (ssGSEA) were performed out to identify the divergent immune cell subtypes and reflect immune cell infiltration. The pathway enrichment analyses about EOPE and LOPE were revealed using gene set variation (GSVA) and ssGSEA. Correlation analyses, performed by Pearson correlation analysis between hub genes, immune cells and enriched pathways, provided details on the underlying molecular mechanisms. Hub genes-miRNA network was performed using the NetworkAnalyst online tool. The identification of placental immunological differences provides a new basis for the development of new potential diagnostic markers and new therapies targeting molecules for preeclampsia.
2 MATERIALS AND METHODS
2.1 The mRNA expression profiling data source
To identify the gene expression datasets, we used the keywords “((early-onset) AND (preeclampsia) AND (late-onset))” and “Expression profiling by array” to search the GEO database. Three GEO datasets (GSE74341, GSE22526 and GSE4707) were screened. These two GSE profiles were disregarded due to the small number of samples and differentially expressed genes examined by GEO2R for GSE22526 and GSE4707. The mRNA expression profiling data was obtained from GSE74341, which was based on GPL16699 Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381. The GSE profile (GSE74341) included seven EOPE, eight LOPE and 10 control patients. GEO is a public database with ethically approved data sources. There are no conflicts of interest or other ethical issues because our study was based on the GEO database. The Immunology Database and Analysis Portal database, which contains 1793 immune-related genes (IRGs), was used to identify IRGs for immunology research. In addition, using the ggplot2 package, version 3.4.4 (https://ggplot2.tidyverse.org), in R software, version 4.2.2 (R Foundation, Vienna, Austria), a principal component analysis (PCA) plot of 25 samples was generated.
2.2 Evaluation of immune status
On GSE74341, estimateR software (https://R-Forge.R-project.org/projects/estimate) and the ESTIMATE method were used to generate the microenvironment score, which includes the immune score and stromal score. Immune scores are defined as level of immune cell infiltration. Stromal scores are defined as degree of stromal cell infiltration.
2.3 Biological function and pathway enrichment analyses
The ClusterProfiler R package, version 4.10.0 (https://bioconductor.org/packages/clusterProfiler), was employed to perform GSEA, Gene Ontology (GO) (http://geneontology.org) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.genome.jp/kegg) pathway enrichment analysis.15 The following criteria were used to evaluate significantly enriched items: false discovery rate (FDR) < 0.25, q value < 0.2 and padj < 0.05. The Molecular Signature Database (MsigDB) was used to download the reference gene list C2.cp.v7.2 symbols.gmt.16 The biological process (BP), cellular component (CC) and molecular function (MF) categories were included in the GO analysis.
2.4 Identification of differentially expressed mRNAs
To analyze differential expression, the limmaR package, version 3.58.1 (https://bioconductor.org/packages/limma), was utilized. Adjusted p and the logarithm of fold change (log2 FC), which represents the effect size of a function, were calculated for each evaluated gene, absolute log2 FC > 1.5 and an adjusted p < 0.05 were defined as significant results. The intersection of the IRGs set and GSE74341 was performed using the UpSetR package, version 1.0.13 (https://CRAN.R-project.org/package=UpSetR), and the overlapping differentially expressed IRGs (DEIRGs) were obtained. The RCircos R package, version 1.2.2 (https://cran.r-project.org/package=RCircos), was employed to analyze the location of DEIRGs on 23 chromosomes. Visual hierarchical cluster analysis with the volcano plot showed the DEIRGs. A heat map of DEIRGs was created using the ComplexHeatmap package in R, version 4.2.1 (https://github.com/jokergoo/complexHeatmap).
2.5 Identification of hub genes
Random forest, a machine learning algorithm, was used to evaluate the relative importance of features.17 Feature importance can help to filter features to a certain extent, thereby filtering hub genes. Using the randomForest package, version 4.7–1.1 (https://cran.r-project.org/package=randomForest), we evaluated the error rate of 1 to 500 trees and selected the number of trees with the lowest error rate. Once the parameters are determined, we construct a random forest tree model. An increase in node purity (IncNodePurity) represents the impact of each variable on the heterogeneity of observations at each node in the classification tree to compare the importance of variables. The larger the value of IncNodePurity, the greater the importance of the variable. The top 20 genes with importance values were identified as DEIRGs of preeclampsia for subsequent analysis. The STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) online database (http://string-db.org) was utilized to create a PPI network of concordant DEIRGs. Then, the visualization of the PPI network based on STRING analysis result was created using Cytoscape, version 3.9.1 (https://cytoscape.org) and core genes were screened by the CytoHubba (a plug-in for Cytoscape, version 3.7.2). Hub genes were acquired through the intersection of RF (top 20), PPICount (top 15), MCC (top 15), Betweenness (top 15) and Degree (top 15).
2.6 Evaluation of immune cell infiltration
CIBERSORTx, a type of deconvolution algorithm with a 1000 permutation count, was performed to evaluate the relative abundance of immune infiltrates, which turned the normalized gene expression matrix into the composition of infiltrating immune cells.18 For a reliable prediction of the immune cell composition, we screened samples with a CIBERSORTx result of p < 0.05. Then, the 22 different types of infiltrating immune cells were visualized by a bar graph constructed using the ggplot2 package, version 3.4.4, and the correlation between immune cell subtypes was represented by a correlation heatmap generated by the corrplot package, version 0.92 (https://github.com/taiyun/corrplot). Violin diagrams were performed to visualize variance analysis of immune cells in EOPE and LOPE samples. The ssGSEA was established using the GSVA package, version 1.50.0 (https://bioconductor.org/packages/GSVA), to quantify the relative immune cell infiltration. We obtained 28 types of immune cells, totaling 782 genes (see Supporting information, Table S1).19 The Vioplot package, version 0.4.0 (https://github.com/TomKellyGenetics/vioplot), was adopted to visualize the immunoscores, analyzed by the Wilcoxon test with two-tailed, to identify differential immune cell subtypes between EOPE and LOPE (p < 0.05).
2.7 GSVA and ssGSEA
GSVA, a non-parametric unsupervised approach, transforms genes from a sample matrix into predefined gene sets without a priori knowledge of experiment design. The variation of pathway activity in each sample was estimated using the KEGG gene sets (c2.cp.kegg.v7.4.symbols.gmt), which were obtained from MSigDB. p < 0.05 and enrichment score change > 1.0 were defined as significantly enriched pathways. Additionally, an enrichment score in each sample, generated by ssGESA, signified the absolute enrichment level of a metagene set within a given dataset. The degree of enrichment of immune-related pathways can be assessed from these results (cytolytic activity, T cell co-inhibition, checkpoint, T cell co-stimulation, antigen-presenting cell [APC] co-stimulation, APC co-inhibition, major histocompatibility complex [MHC] class-1,type-1 interferon [IFN] response, inflammation-promoting, para-inflammation, human leukocyte antigen [HLA], type-2 IFN response and cytokine-cytokine receptor interaction [CCR] in current immunology research20, 21).The metagene set was shown in the Supporting information (Table S2).
2.8 Pearson correlation analysis
The association between hub genes, immune cells and crucial molecular pathways was examined using Spearman correlation analysis to shed light on the putative molecular process behind the onset and progression of preeclampsia. p < 0.05 was considered statistically significant.
2.9 miRNA prediction of hub genes and miRNA-mRNA construction
The miRNA of five hub genes were predicted from NetworkAnalyst (http://www.networkanalyst.ca/faces/home.xhtml) using a score ≥ 0.95 as the cutoff criterion. Cytoscape was utilized to construct a miRNA-hub gene regulatory network. FunRich software 3.1.3 (http://funrich.org) was used to predict the biological pathways, potential transcription factors and molecular function of miRNAs.22
2.10 Clinical samples and qRT-PCR
Placental samples were collected from 40 pregnant women, including 20 EOPE patients and 20 LOPE patients. The present study (KY-2020-007) was permitted by the ethics committee of the Women and Children's Hospital of Xiamen university and all participants provided written informed consent. Preeclampsia was diagnosed based on the guideline of International Society for the Study of Hypertension (ISSHP).23 Placental tissues were obtained from areas without obvious infarcts/necrosis within 20 min after delivery. Then placental samples were rapid frozen by liquid nitrogen and stored at −80°C.
Trizol (Invitrogen, Waltham, MA, USA) was used to extract total RNA from placental samples. Reverse transcription of RNA using ABScript II RT Master Mix for qPCR with gDNA Remover (Abclonal, Woburn, MA, USA). qPCR was performed using 2X Universal SYBR Green Fast qPCR Mix (ABclonal). The clinical characteristics of all participants and gene-specific PCR primers are provided in the Supporting information (Tables S3 and S4).
2.11 Statistical analysis
The data were analyzed using R software, version 4.2.2. All statistical analyses are expressed as the mean ± SD and significance was tested using the Student's t-test. p < 0.05 was considered statistically significant.
3 RESULTS
3.1 Evaluation of immune status
As shown in Figure 1A, PCA plot of 25 samples indicated that the repeatability of data from GSE74341 was satisfactory. To evaluate the intragroup data standardization level, Figure 1B shows that the black lines are generally on the same straight line and the results demonstrated that the standardization level of data from GSE74341 was also satisfactory. Then, the immune status in GSE74341 was evaluated using the ESTIMATE algorithm. Immune score and stromal score of EOPE were significantly lower compared to LOPE and normal samples (Figure 1C–D). The immune scores and stromal scores did not differ significantly between LOPE and normal samples. The results demonstrated that immune elements of the placenta play a major role in the etiopathogenesis of EOPE.
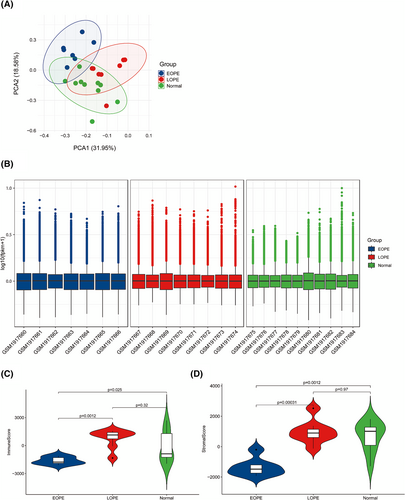
3.2 The involvement of the immune system during EOPE
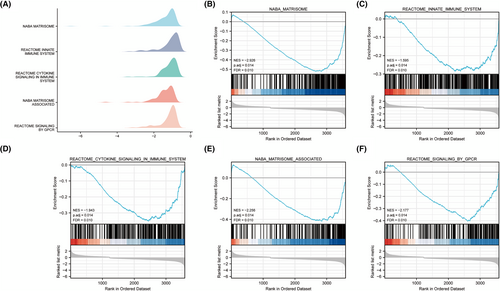
We performed GSEA to further explored immune system in EOPE and LOPE, and the gene expression data in GSE74341 of EOPE and LOPE samples were analyzed holistically. The results showed that immune system-related genes were primarily downregulated and extremely enriched at a nominal padj < 0.05 in EOPE samples (Figure 2A–F). GSEA indicated the involvement of immune responses during EOPE.
3.3 DEIRGs and functional enrichment analysis
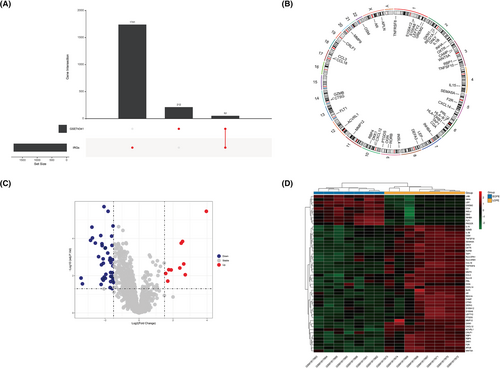
Two hundred sixty-four DEGs were identified in the GSE74341 series (see Supporting information, Table S5) and the UpSet plot showed 52 overlapping genes (DEIRGs) in GSE74341 series and IRGs set (Figure 3A). Figure 3B shown the chromosomal location of the 52 DEIRGs. The volcano plot shows the 52 DEIRGs, including 10 upregulated and 42 downregulated genes (Figure 3C). The gene expression of the 52 DEIRGs is shown in Figure 3D.
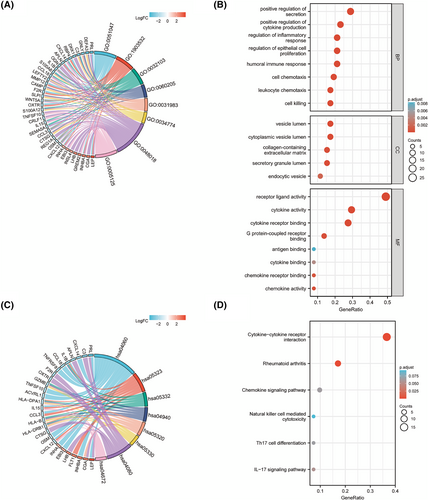
Fifty-two DEIRGs were subjected to GO and KEGG enrichment analysis. The result showed that these DEIRGs were corrected to immune-related functions. We performed the GO term enrichment analysis for 52 DEIRGs and Figure 4A,B showsd the top significant clusters of enriched sets, such as cytokine activity, chemokine activity, humoral immune response, regulation of inflammatory response, cell chemotaxis and cell killing. The KEGG biological pathway enrichment analysis of 52 DEIRGs showed CCR, rheumatoid arthritis chemokine signaling pathway, interleukin (IL)-17 signaling pathway, T helper (Th) 17 cell differentiation and natural killer (NK) cell-mediated cytotoxicity mainly involved (Figure 4C,D). Detailed data and further details of the enrichment analysis are provided in the Supporting information (Table S6). The results show that pro-inflammatory factors and pathways play crucial roles in the pathological process between EOPE and LOPE.
3.4 Identification of hub genes
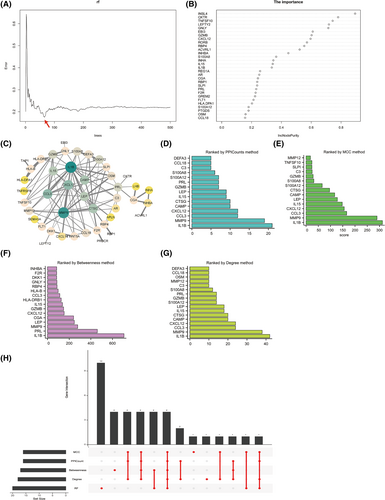
To screen out persuasive core genes, we performed RF for feature selection and accurately estimated the importance of each feature. The results showed that the model error was minimum when the number of trees for regression was 66 (Figure 5A). As shown in Figure 5B, the corresponding model, built with a sorting diagram, indicated the importance of variables-DEIRGs. The visualization of the PPI network of 52 DEIRGs was constructed using Cytoscape (Figure 5C). Fifty-two nodes (genes) and 113 edges (interactions) were identified in the PPI network with a enrichment p < 1.0 × 10–16. The number of nodes for each gene (PPICount) was calculated by utilizing the STRING online database (Figure 5D). To identify hub genes, Cytoscape's cytoHubba plug-in was utilized. We used three commonly classification methods (MCC, Betweenness and Degree) in cytoHubba and these results are shown in Figure 5E–G. We applied the six methods mentioned above, which represent aspects of machine learning algorithm and biological function, to acquire four hub genes through intersection (IL15, GZMB, IL1B and CXCL12) (Figure 5H). With a combination RF and PPI network, the four hub genes may be correlated with EOPE progression.
3.5 Immune cell infiltration analysis
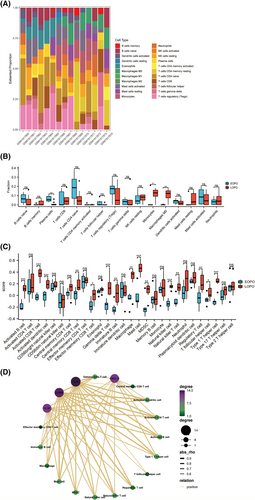
Microarray data from GSE74341 was subjected to CIBERSORTx analysis to identify the proportion of 22 immune cell types aiming to explore the distinct immune landscape in EOPE and LOPE. In Figure 6A, stacked bar plots shown the abundance of 22 immune cell types. The fraction of 22 immune cell types in the samples of EOPE and LOPE is shown by the boxplots (Figures 6B). Compared with LOPE, monocytes (p < 0.05) and macrophages M0 (p < 0.01) are significantly lower in EOPE, whereas plasma cells (p < 0.01) is relatively higher. This finding indicates that monocytes and macrophages M0 were significantly decreased in EOPE. To further quantify the enrichment score of immune cell types, we use ssGSEA. The score of activated B cell, activated CD8 T cell, activated dendritic cell, central memory CD8 T cell, effector memory CD8 T cell, gamma delta T cell, immature B cell, macrophage, mast cell, myeloid-derived suppressor cell, NK cell, NK T cell, regulatory T cell (Treg), T follicular helper cell and type 1 Th cell were significantly lower in EOPE than those in LOPE samples (Figure 6C). Spearman correlation analysis was performed to investigate whether the four hub genes were associated with immune infiltration in EOPE. Based on enrichment score of ssGSVA, our hub genes were significantly related to activated B cell, activated CD8 T cell, Treg, macrophage, NK cell, NK T cell and type 1 Th cell (Figure 6D). The innate and adaptive maternal immune cells were significantly decreased in EOPE compared to LOPE.
3.6 GSVA and ssGSEA
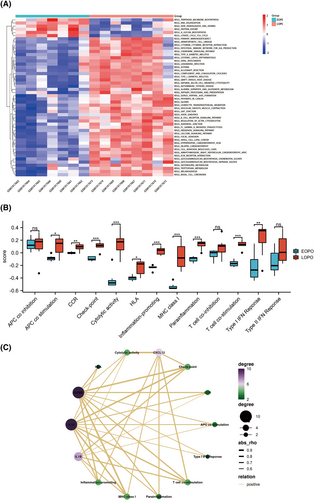
GSVA was utilized to investigate for biological differences between EOPE and LOPE in GSEM databases. The results showed that (primary immunodeficiency, CCR, chemokine signaling pathway and NK cell-mediated cytotoxicity) were significantly deficient in EOPE samples compared to LOPE (Figure 7A). Additionally, ssGSEA was performed to screen predefine immune-related pathways in EOPE. The results showed that APC co-stimulation, CCR, checkpoint, cytolytic activity, HLA, inflammation-promoting, MHC class I, para-inflammation, T cell co-stimulation and type I IFN response were involved in the mechanism of EOPE (Figure 7B). The association between hub genes and enriched molecular pathways was identified by Pearson correlation analysis to reveal the molecular pathways that may be involved in EOPE. Hub genes were strongly associated with CCR, para-inflammation, MHC class I and T cell co-stimulation based on the enrichment score of ssGSVA (Figure 7C).
3.7 Validation of hub genes
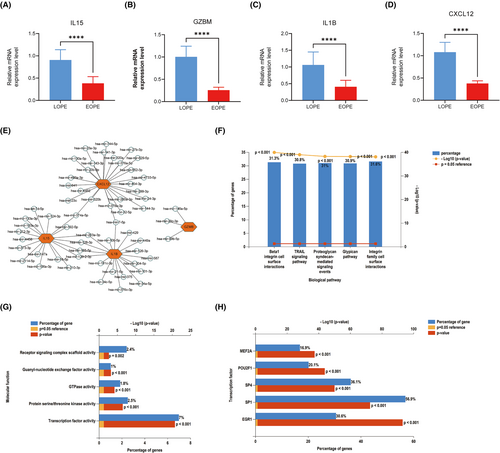
qRT-PCR was used to verify the expression of the four hub genes in 40 patient samples. Compared with placenta of LOPE, IL15, GZMB, IL1B and CXCL12 were significantly downregulated in EOPE (Figure 8A–D).
3.8 Further miRNAs prediction of hub genes
To further predict miRNAs of hub genes, we established a miRNA–hub gene regulatory network. The result showed that hsa-miR-374a-5p, hsa-miR-203a-3p, hsa-miR-128-3p, hsa-miR-155-3p, hsa-miR-7-5p and hsa-miR-129-2-3p had higher amounts of cross-linked genes (≥ 2) (Figure 8E). Afterwards, 74 miRNAs were submitted to FunRich. The enriched biological pathways, according to the results of the enrichment analysis, included beta1 integrin cell surface interactions, TRAIL signaling pathway, proteglycan syndecan-mediated signaling events, glypican pathway and integrin family cell surface interactions (Figure 8F). The molecular functions were significantly enriched in guanyl-nucleotide exchange factor activity, Protein serine/threonine kinase active, receptor signaling complex scaffold activity, GTPase activity and transcription factor (Figure 8G). As shown in Figure 8H, the top five transcription factors of these miRNAs were EGR1, SP1, SP4, POU2F1 and MEF2A.
4 DISCUSSION
Despite the understanding of preeclampsia becoming clear with more and more studies on the mechanism, there are still numerous key questions waiting to be answered. Recent genome-wide association researches and immunological studies have focused on the correlation between immune maladaptation and preeclampsia.24-26 However, the different pathogenesis of the main two preeclampsia subtypes, EOPE and LOPE, is usually ignored. The placental immune process is crucial for maintaining a tolerogenic environment and the development of the placenta and fetus.27, 28 Although EOPE and LOPE share some common pathogenic mechanisms such as systemic maternal inflammation, the changes in the placental immune system remain incompletely explained. Here, we explored the role of immune regulation in preeclampsia, particularly in EOPE and LOPE. Through analyzing placental RNA profiles from EOPE, LOPE and normal pregnancy, we indicated that the immune status was impaired in EOPE samples, but not in LOPE samples. Moreover, we identified that IL15, GZMB, IL1B and CXCL12 were the hub genes regulating immune dysfunction in EOPE. These hub genes potentially cause immune dysregulation by modulating cytokine-cytokine receptor, para-inflammation, MHC I and T cell co-stimulation pathways, thereby contributing to the pathogenesis of EOPE, which could be regulated by hsa-miR-374a-5p, hsa-miR-203a-3p, hsa-miR-128-3p, hsa-miR-155-3p, hsa-miR-7-5p and hsa-miR-129-2-3p. Additionally, the aforementioned hub genes were further validated in the clinical samples.
Preeclampsia, especially EOPE, was considered to be secondary to abnormal uterine spiral artery remodeling and insufficient placental perfusion.29 The infiltration of placental immune cells is closely related to the remodeling of spiral arteries. Disorders in these processes may cause or lead to preeclampsia. In the present study, we analyzed GSE75010 datasets which contained EOPE, LOPE and normal samples. The results, consistent with previous reports,30 showed that the immune status was impaired in EOPE samples, but not in LOPE samples. To further explore the pathogenesis of the two preeclampsia subtypes, we focused on placental immune alterations between EOPE and LOPE. According to GSEA analysis I, innate immunity and cytokine signaling in the immune system are the most likely immunological components implicated in the pathogenesis and immunoregulatory mechanisms of EOPE. We screened 52 DEIRGs between EOPE and LOPE samples. The PPI network (cytoHubba), representing biological function, and RF, representing the machine learning algorithm, were used to reveal four hub genes (IL15, GZMB, IL1B and CXCL12). Importantly, qRT-PCR results confirmed that EOPE samples had considerably lower expression levels of IL15, GZMB, IL1B, and CXCL12 than LOPE samples. A CIBERSORTx examination of immune infiltration revealed immune cells, such as monocytes and M0 macrophages, which are much less abundant in EOPE. Notably, ssGSEA showed that hub genes were significantly correlated with activated B cells, activated CD8 T cells, Tregs, macrophages, NK cells, NK T cells and type 1 Th cells. According to GSVA and ssGSEA, hub genes were significantly related to CCR, checkpoint, APC co-inhibition and type I IFN response. Finally, to further predict miRNAs of hub genes, we established a miRNA–hub gene regulatory network. The results showed that hsa-miR-374a-5p,hsa-miR-203a-3p,hsa-miR-128-3p, hsa-miR-155-3p, hsa-miR-7-5p and hsa-miR-129-2-3p had higher amounts of cross-linked genes (≥ 2).
From the perspective of etiology, EOPE and LOPE have different etiological features. LOPE is more related to maternal factors, while EOPE is more correlated to placental factors.31 Biomarkers (IRGs) have been identified with remarkable accuracy and rapidity for disease surveillance and treatment with the introduction of large-scale, public and high-throughput genomic assays in recent years.32 Analyzing related characteristics in EOPE and LOPE may contribute to explore the pathogenesis and treatment of preeclampsia. The investigation of relevant immune pathways and immune responses in diverse tissue types is performed using an approach that has arisen in the field of oncology that makes use of the expression of IRGs. Even with conditions such as pancreatic cancer that have an immune-suppressive milieu, this technique's great sensitivity enables the detection of non-abundant immune cells.33, 34 Therefore, we investigated the immune profile of preeclampsia based on this technique. To our knowledge, this is the first study to report the IRG signature and immune infiltration features between EOPE and LOPE. Our study may contribute to the identification of novel biomarkers for preeclampsia and reveal the prospective pathogenesis and potential therapeutic targets of preeclampsia.
The coordination of the innate and adaptive immunity in maternal immune system is crucial for a successful pregnancy. The maternal immune system must balance the tolerance of the allogeneic fetus and placenta and the protection of the fetus against pathogens. The interaction between innate immune cells and other cells plays a crucial role in normal pregnancy.10 Most of innate immune cells in the placenta are macrophages and NK cells, which may comprise about 90% of the total immune cell count.35 In the present study, immune infiltration analysis based on CIBERSORTx and ssGSEA showed that macrophages, NK cells and mast cells are apparently absent in EOPE samples, compared with LOPE samples, which may reveal a compromised innate immune system in EOPE. Numerous studies focused on the functions of macrophages in human placentas and revealed the various roles of these cells during pregnancy. They can particularly be found around spiral arteries that show disorganization and disruption of endothelial cells and vascular smooth muscle cells, while no extravillous trophoblast display at that time.36 Meanwhile, decidual macrophages found in the vicinity of spiral uterine arteries secrete angiogenic molecules.37 This indicates that macrophages may play an important role in the preparation of spiral arteries for following remodeling by trophoblast cells. Apoptotic trophoblast cells can be observed during spiral artery remodeling by trophoblast cells.38 Macrophages in the vicinity of spiral arteries at that time may have an essential function in engulfing these apoptotic trophoblast cells, thus blocking the release of pro-inflammatory compounds into the decidua.39 The results of CIBERSORTx analysis also showed that monocyte counts decreased in EOPE samples. It is widely identified and accepted that monocytes are the exclusive source of tissue macrophages; however, whether macrophages in the placental bed originate from monocytes during pregnancy remains uncertain.40 It is worth examining further whether there are any links in the absence of macrophages and monocytes in the EOPE placenta. As an important role in the remodeling of spiral arteries, NK cells secrete cytokines (mainly macrophage colony-stimulating factor 1 and TGF-B1), regulate trophoblast invasion and recruit macrophages to participate in vascular remodeling.41 NK cells are implicated in pregnancy complications associated with poor placentation, such as preeclampsia. However, the importance of NK cells in the pathogenesis of preeclampsia remains some controversy because multiple studies have provided conflicting data. Numerous studies have revealed increased frequencies or numbers of NK cell in reproductive and gestational tissues of women with preeclampsia,42, 43 whereas other studies have revealed a diminished numbers or frequencies in preeclampsia compared to those tissues of normal pregnancies.44 Different pathological processes of various preeclampsia subtypes, alterations in specific NK cell phenotypes and various evaluating techniques of immune cell populations may contribute to such discrepancies. In the present study, based on the expression of IRGs from expression profiling using arrays, we found that NK cell counts are significantly reduced in EOPE placenta compared to those in LOPE placenta, which may provide a reasonable basis for elucidating the roles of NK cells in preeclampsia. A lower abundance of mast cells in the placenta of EOPE is in accord with previous studies.30 However, such lower abundance changes of mast cells rarely present in other subtypes of preeclampsia. Mast cells secrete angiogenic factors,45 and their depletion leads to abnormal spiral artery remodeling. Studies have confirmed that an increase in the number of mast cells and an increase in microvessel density can be observed in the placenta of patients with gestational diabetes. These limited reports indicate the potential involvement of mast cells in the pathogenesis of preeclampsia. Thereby, the involvement of mast cells may be demonstrated by mechanistic studies.
Furthermore, we have demonstrated the alteration of adaptive immune cells at the maternal-fetal interface with EOPE and LOPE. Immune infiltration analysis based on ssGSEA found that adaptive immune cells, such as Tregs, T follicular helper cells and Type 1 T helper cells, are apparently absent in EOPE samples compared to LOPE samples. Tregs are conceived to be arguably the most important role in the maintenance of maternal tolerance to placental antigens and fetal.46 Studies revealed systemically increasing of Tregs in normal pregnancy and present locally of those cells in the human decidua.47 Numerous studies have consistently shown that the preeclampsia placenta has less Tregs than the normal placenta.48 However, one study did not report such a reduction. The EOPE placenta has a more severe absence of Tregs compared to EOPE,48 which also suggested differences in the pathophysiology of the two subtypes. A reduction of the number of Tregs is mirrored by a concomitant increase in the number of effector T cells, particularly T helper 17 cells, although we did not observe such an increase. In the present study, gamma delta T cell, effector memory CD8 T cells, central memory CD8 T cells and activated CD8 T cell were more abundant in LOPE samples compared to EOPE samples. Why this happens is an intriguing question. Gamma delta T cells are composed of T cells that express a TCR composed of a γ-chain and a δ-chain. These cells, which maintain high levels throughout pregnancy, highly express TLRs and can be either cytotoxic or regulatory depending on the abundance of TLR activation and cytokines.49 The investigation of such gamma delta T cells in the pathogenesis of preeclampsia has been limited, but merits attention and investigation. However, one study revealed that pregnant mice with activated TLR3 and TLR7 exhibit inflammation and preeclampsia-like characteristics, and either genetic or pharmacological reduction of gamma delta T cells may prevent pregnancy-related hypertension.50 The increased infiltration of CD8+ T cells is an immunological response. Some studies reported increased levels of placental CD8+ T cells in pregnancy complication with preeclampsia or chorioamnionitis. Placental oxidative stress, which could contribute to T cell infiltration, is a feature of preeclampsia. Hypoxia causes an upregulation of migration inhibitory factor levels, which attracts T cells to the human placenta in vitro.51 However, in villitis of unknown etiology, T cell infiltration is higher in LOPE than in normal pregnancy but not in EOPE,52 consistent with our results. This may further explain the difference in pathogenesis between EOPE and LOPE.
IL-15 plays important roles in regulating activation and proliferation of immune cells. Studies indicated that IL-15 expression was elevated in the serum of patients with preeclampsia,53 whereas its expression was reduced in the placentas.54 GZMB participates in modulating cytokine release, extracellular matrix proteins degradation, and so on. At present, studies regarding the function of GZMB in preeclampsia are scarce. IL1B is a cytokines produced by immune cells, involving in diverse cellular process such as cell proliferation, differentiation and so on. Research has revealed that elevated S100A9 could promote IL1B secretion from the placenta and trophoblast in preeclampsia.55 CXCL12 regulates a variety of cellular functions including immune surveillance, inflammation response and so on. Expression of CXCL12 in severe preeclamptic placentas was significantly downregulated compared to that in mild preeclampsia and normal pregnancy.56 However, roles of the hub genes in EOPE remain unclear. Here, we found that the expression of IL-15, GZMB, IL1B and CXCL12 was downregulated in the placentas of EOPE compared to LOPE, indicating the potentially roles in the development of EOPE.
GSVA results showed several classic and common signaling pathways, as inflammatory mediators and modulate immune responses, including primary immunodeficiency, CCR, NK cell-mediated cytotoxicity and the chemokine signaling pathway, were deficient in EOPE samples. SsGSEA results demonstrated that CCR, para-inflammation, MHC class I and T cell co-stimulation were significantly deficient in EOPE samples and their enrichment scores significantly correlated with hub genes. It has been indicated that CCR involved in modulating PE onset,57, 58 including early-onset HELLP syndrome59 and early preeclampsia,60 may be associated with mechanisms of regulating immune responses such as macrophages.61 Furthermore, immune cells could contribute to preeclampsia pathogenesis through modulating inflammatory responses.58MHC class I plays an essential role in maternal-fetal immune balance.10, 62 Additionally, T cells are crucial adaptive immune cells during pregnancy, and their abnormal activation is closely related to preeclampsia onset.10, 28 Here, we also indicated that CCR, para-inflammation, MHC class I and T cell co-stimulation probably are the main immune pathways that contribute to pathological process of EOPE. However, additional research is needed to verify the specific underlying mechanism.
In the present study, there are some limitations. First, all of the analysis was based on public databases and not all functional assays were conducted. In addition, the sample size contained in public databases was relatively small. As a supplement, to demonstrate the robustness of the results regarding hub genes, we collected 20 placental samples from EOPE and 20 placental samples from LOPE, and performed qRT-PCR. The results obtained were consistent with the analysis results of the database. Second, the immune cell infiltration assessment realized by CIBERSORTx provides relative proportions and may not be reflective of absolute infiltration for inter-sample comparisons.63 Strictly speaking, additional studies will be essential to quantify the potential changes in absolute immune infiltration that may affect preeclampsia.
5 CONCLUSIONS
Once the molecular mechanisms and potential therapeutic targets that cause placental dysfunction have been intensively studied, this may provide significant assistance for early diagnosis of preeclampsia. Interactions between the mother and the placenta are significantly influenced by immune cells. The comparison of immune-related genes signature and immune infiltration features in early- and late-onset preeclampsia may help to explain the cause of placental dysfunction. The present study revealed that the innate and adaptive immune systems are severely impaired in placentas of EOPE, compared to LOPE, as reflected by downregulated immune system pathways, and a lower abundance of innate and adaptive immune cells. Four IRGs (IL15, GZMB, IL1B and CXCL12) were tightly linked to the pathogenesis of EOPE, probably by effecting CCR, para-inflammation, MHC class I and the T cell co-stimulation pathways involved in the onset of EOPE. This study may provide a new basis for discriminating between EOPE and LOPE and acknowledging the role of the immune landscape in the eventual interference and tailored treatment of EOPE.
AUTHOR CONTRIBUTIONS
XZ was responsible for conceptualization. WW was responsible for data curation. WW was responsible for formal analysis. QW was responsible for funding acquisition. HL was responsible for investigations. HL, WW and XL were responsible for methodology. HL, XL and MY were responsible for project administration. WY was responsible for resources. WY and XY were responsible for software. WY was responsible for supervision. XY was responsible for validation. QW was responsible for visualization. QW and XY were responsible for writing the original draft. QW and XY were responsible for reviewing and editing.
ACKNOWLEDGMENTS
This work was supported by Xiamen Key Laboratory of basic and clinical research on major obstetric diseases, Department of Obstetrics, Women and Children's Hospital, School of Medicine, Xiamen University. We are indebted to all the patients who participated in studies of GEO databases. We gratefully acknowledge the financial supports by Xiamen Natural Science Foundation Project under Grant number 3502Z20227142, as well as Xiamen Health Care projects number 3502Z20224ZD1221.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
The article and the Supporting information contain the original contributions presented in this study; further requests can be made to the corresponding author.




