Expression profile of lncRNAs and miRNAs in esophageal cancer: Implications in diagnosis, prognosis, and therapeutic response
Abstract
Esophageal cancer is the seventh most common cancer worldwide. Although a number of environmental and lifestyle-related risk factors have been identified for this kind of cancer, the exact molecular mechanisms of tumor evolution have not been clarified yet. Long noncoding RNAs (lncRNAs) and microRNAs (miRNAs) as important regulators of gene expression and chromatin configuration have essential roles in the pathogenesis of esophageal cancer. They have been shown to alter the function of cancer-related signaling pathways such as phosphoinositide 3-kinase/protein kinase B and Wnt pathway, thus they might modulate the response of patients to pathway-targeted therapies. Moreover, a number of lncRNAs, such as AFAP1-AS1, UCA1, HOTAIR, LOC285194, and TUSC7, are involved in conferring chemoresistant/radioresistant in esophageal cancer cells. A complex network of interaction exists between lncRNAs and miRNAs in the context of esophageal cancer. Finally, various panels of lncRNAs and miRNAs have been introduced that can predict the survival of esophageal cancer patients. In this review article, we summarize the recent findings regarding the role of miRNAs and lncRNAs in the pathogenesis of esophageal cancer with the special focus on their regulatory roles on signaling pathways, their potential as diagnostic/prognostic markers, and their relevance with therapeutic response.
1 INTRODUCTION
Esophageal cancer accounts for 572,000 new cases of cancer which put this type of cancer at the seventh position with regard to incidence. Moreover, it ranks sixth among cancers in mortality rate (Bray et al., 2018). There is a sex bias in both incidence and mortality of esophageal cancer in a way that more than two-thirds of cases occur in males and mortality is twofold to threefold higher in this sex (Bray et al., 2018). Histological examination has classified esophageal cancer into two main categories, namely esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC) with the former being more prevalent in South East and Central Asia, while the latter is common in western countries (Arnold, Soerjomataram, Ferlay, & Forman, 2015). Differences in the distribution of esophageal cancer subtypes have been attributed to the presence of environmental factors (Watanabe, 2015). Among possible risk factors for ESCC are alcohol drinking, smoking, betel quid chewing, and drinking very hot beverages with the first two factors having synergic effects (Thun, Linet, Cerhan, Haiman, & Schottenfeld, 2017). Meanwhile, obesity and gastroesophageal reflux disease are regarded as risk factors for EAC (Thun et al., 2017). Despite the improvements in diagnostic and therapeutic options, the overall survival of patients with esophageal cancer is still poor and is not much higher than 40% for 5 years (Hou et al., 2019). On the basis of the observed close association between stage at diagnosis and survival rate (Hou et al., 2019), identification of novel diagnostic markers for this kind of cancer has practical significance.
Recent studies have shown aberrant expression of a number of long noncoding RNAs (lncRNAs) in esophageal cancer (Qiao, Li, Zhao, Yan, & Sun, 2018). These transcripts comprise the vast majority of noncoding sequences of the human genome and are involved in biological processes, such as imprinting, chromatin remodeling, cell cycle regulation, and cell differentiation (Kanduri, 2016). All of these processes are potentially involved in the pathogenesis of human cancers, yet participation of lncRNAs in carcinogenesis process is not limited to their regulatory functions on these processes. LncRNAs regulate many cancer-associated signaling pathways, such as phosphoinositide 3-kinase/protein kinase B (PI3K/AKT), hypoxia-inducible factor 1-α (HIF-1α), and Hippo signaling pathways (P.-F. Fu, Zheng, Fan, & Lin, 2019). LncRNAs regulate gene expression at different levels through recruitment of chromatin-modifying proteins, binding with transcription factors, and decreasing their bioavailability (K. C. Wang & Chang, 2011), acting as decoys for microRNAs (miRNAs) and enhancing expression of miRNA target genes (F. Xu & Zhang, 2017), and modulating subcellular localization and function of proteins (Noh, Kim, McClusky, Abdelmohsen, & Gorospe, 2018). miRNAs are also involved in the pathogenesis of esophageal cancer. These short noncoding RNAs regulate expression of target genes at posttranscriptional level mostly through binding to a 3′-untranslated region of messenger RNAs (mRNAs). Vast regulatory functions of miRNAs have potentiated them as predictive markers for diagnosis and prognosis of different cancers including esophageal cancer (Vrana et al., 2017). In the current review article, we summarize the recent findings regarding the role of lncRNAs and miRNAs in the pathogenesis of esophageal cancer with the special focus on their regulatory roles on signaling pathways and their potential as diagnostic/prognostic markers and therapeutic targets.
2 EXPRESSION PROFILE OF LncRNAs IN ESOPHAGEAL CANCER
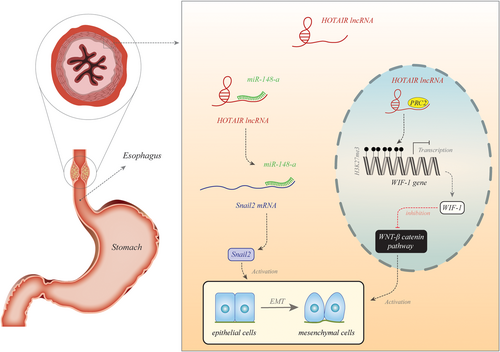
Several independent groups have assessed the expression of lncRNAs in esophageal cancer samples or serum samples of patients. On the basis of the results of these studies, we can classify these transcripts to oncogenic and tumor suppressor transcripts. HOTAIR is a representative of oncogenic lncRNAs. Figure 1 shows the molecular mechanism of its involvement in esophageal cancer.

Tables 1 and 2 show up- and downregulated lncRNAs in the esophageal cancer samples, respectively.
| lncRNA | Numbers of clinical samples | Assessed cell line | Targets/regulators | Signaling pathways | Function | Patient's prognosis | Reference |
|---|---|---|---|---|---|---|---|
| DUXAP8 | 78 Pairs of esophageal cancer tissues and normal adjacent tissues | TE-1, ESCCA109, ESCCA9706, KYSE520, HEESCC | – | Wnt/β-catenin | Since its knockdown can inhibit cell proliferation and invasion, DUXAP8 may promote tumor development | Correlate with poor prognosis | Xu et al. (2018) |
| NEAT1 | 96 Patients with ESCC | SHEE, SHEEC, KYSE150, KYSE30, KYSE70, KYSE140 | – | – | By improving tumor cell migration and invasion, may play an important role in disease progression | Predicts poor prognosis | Chen, Kong, Ma, Gao, and Feng (2015) |
| linc00460 | 65 Patients with ESCC | EC109, KYSE150, KYSE450, Het-1A | CBP/P300 | – | By acting as an oncogene may be used as a diagnostic and therapeutic biomarker | Poor prognosis | Liang et al. (2017) |
| HOTAIR | 100 Patients with ESCC and 56 adjacent nonneoplastic controls | TE1, TE3, TE7, TE8, KYSE30, KYSE180, KYSE150, KYSE140, KYSE510, KYSE450, BIC1 | – | – | Plays an important role in ESCC progression via gene regulation and may serve as an epigenetic target for the treatment of patients | Shorter survival time | X. Li et al. (2013) |
| HOTAIR | 78 ESCC specimens | KYSE30, KYSE150, KYSE450, KYSE510, KYSE180 | – | – | It shows an important role in the development and progression of patients with ESCC and provides a novel therapeutic target | Shorter survival time | F. J. Chen et al. (2013) |
| HOTAIR | 137 ESCC tissues and adjacent normal samples | KYSE30, KYSE140, KYSE180, KYSE410, KYSE510 | WIF-1 | Wnt/β-catenin | It can improve ESCC cell metastasis and act as a novel therapeutic target | Poor prognosis | Ge et al. (2013) |
| HOTAIR | 93 ESCC samples and adjacent normal tissues | – | – | – | By playing an important role in disease progression, it can be used as a potential biomarker | Poor OS | F. J. Chen et al. (2013) |
| HOTAIR | 40 Paired of tumor and adjacent tissues | KYSE30, KYSE150, TE-1, Eca-1, Eca-109, HEEpiC | miR-148a | – | Having an important role in the development and progression of ESCC may be useful in therapeutic approaches | Poor OS | F. Xu and Zhang (2017) |
| HOTAIR | Serum samples from 50 patients with ESCC and 20 healthy subjects | – | – | – | Serum HOTAIR can be a putative biomarker for the diagnosis of ESCC | – | Wang et al. (2017) |
| HOTTIP | 78 Paired of cancerous and the adjacent noncancerous tissues | EC109, EC9706, KYSE30, KYSE450 | – | EMT pathway | Acting as an oncogene by improving cell proliferation and migration, it may serve as a new target for therapy of patients with ESCC | – | Chen et al. (2016) |
| ANRIL | 8 Patients with ESCC and adjacent nontumor controls | TE1, ECA109 | – | TGF-β /Smad | It has an influence on ESCC development and cell proliferation | – | D. Chen et al. (2014) |
| XIST | 78 Patients with ES | HEEC, SKGT-4 and Bic-1, TE-1, HCE-4, HCE-7 | miR-494 | JAK2/STAT3 | By playing as an oncogene it may develop esophageal carcinoma | – | Z. Chen et al. (2019) |
| SOX2OT | 66 Patients with ESCC and paired nontumor tissues | KYSE450, KYSE150, EC109, EC9706 | – | – | It can develop ESCC growth | – | Y. Wu et al. (2018) |
| NORAD | 106 Patients with ESCC | – | – | – | Playing as a molecular predictor of poor prognosis and can be seen as a target for therapy | Poor DFS and OS | X. Wu et al. (2017) |
| PCAT-1 | 75 ESCC tissues and 75 adjacent normal samples | – | – | – | It can be important in ESCC pathogenesis and serve as a prognostic biomarker | – | Razavi and Ghorbian (2019) |
| PCAT-1 | 130 Cancerous tissues and adjacent noncancerous patients | – | – | – | Correlated with tumor invasion and metastasis so may be useful in the treatment of patients with ESCC | Poor prognosis | W.-H. Shi et al. (2015) |
| MALAT1 | 132 Patients with ES | EC109, KYSE150, HEF-1A | – | – | It can be seen as a prognosis predictor | Shorter OS | C. Huang, Yu, Yang, and Lin (2016) |
| MALAT1 | 106 Paired of ESCC tissues and adjacent noncancerous samples | TE1, TE7, EC1, EC109, KYSE70, KYSE450 | β-catenin | – | Associates with tumor proliferation, migration, and formation so might be used as a potential target for ESCC | Poor OS | Wang et al. (2016) |
| UCA1 | 90 ESCC samples and adjacent nontumor tissues | EC109, EC9706, KYSE150, KYSE510, NE1 | – | – | Have a role in proliferation, migration, and invasion ability of malignant cells so can be used as a prognostic/diagnostic biomarker | Poor prognosis | J.-Y. Li, Ma, and Zhang (2014) |
| UCA1 | 66 Patients with ESCC and adjacent normal tissues | EC9706, KYSE | Sox4 | – | Promotes cell proliferation and can act as a therapeutic target for patients with ESCC | Poor prognosis | Jiao et al. (2016) |
| FOXCUT | 82 ESCC tissues and adjacent noncancerous samples | KYSE30, KYSE70, KYSE140, KYSE150, KYSE180 | – | – | Involves in tumor progression and survival of cancerous cells, so can be seen as a biomarker for prognosis and targeted therapy | Poor prognosis | F. Pan et al. (2014) |
| ZEB1-AS1 | 87 Paired of ESCC samples | – | – | – | Associates with tumor progression and uses as an independent prognostic factor | Poorer OS and DFS | Y.-L. Wang, Bai, Yao, Guo, and Wang (2015) |
| LINC01234 | – | KYSE-450, EC-109, CEC2, EC-9706, KYSE-180 | – | – | Can affect migration, invasion, proliferation and apoptosis of cancerous cells | – | Ghaffar et al. (2018) |
| TUG1 | 218 Paired of ESCC and adjacent normal tissues | – | – | – | Related to chemotherapy resistance so may be used as a potential target for therapy of patients with ESCC | Poor prognosis | L. Jiang et al. (2016) |
| DANCR | 32 Patients with ESCC | ECA109, TE-1 | – | – | Promotes proliferation, migration and invasion of tumor cells | – | H. Shi et al. (2018) |
| BANCR | 142 Patients with ESCC and adjacent normal tissues | KYSE-30, KYSE-70, KYSE-140, KYSE-150, KYSE-450, KYSE-510, TE-10, TE-12, Het-1A | – | – | Acting as a novel tumor biomarker, it can be used as a potential therapeutic/prognostic factor | Poor prognosis | Z. Liu, Yang, Xu, and Cao et al. (2016) |
| SBF2-AS1 | 51 Paired of ESCC samples | ECA-109, HEEC, TE-1, KSYE-410 | – | – | Having an important role in proliferation and invasion of tumor cells and may be used as a therapeutic target for patients | Poor prognosis | R. Chen et al. (2018) |
| TP73-AS1 | 60 ESCC tissue samples and the adjacent normal tissues | HET-1A, Eca109, EC-1, EC9706, KYSE30, KYSE150 | – | – | Acts as an important biomarker for prognosis and diagnosis of patients | – | Zang et al. (2016) |
| PVT1 | 104 Patients with ESCC | HEK293T, KYSE30, KYSE410, KYSE520, KYSE510, KYSE140, KYSE150, Eca109, Eca9706, NE1 | miR203/LASP1 | – | Can promote tumor progression via PVT1/miR-203/LASP1 axis in patients with ESCC | Poor prognosis | X. Wu et al. (2017) |
| CASC9 | 44 ESCC and corresponding normal tissues | TE1, Kyse150, EC109, EC9706, and EC1, 293T | – | – | Having an important role in migration and invasion, it may serve as a potential therapeutic target for patients with ESCC | Poor prognosis | Z. Pan et al. (2016) |
| PlncRNA-1 | 73 Paired of ESCC and their matched normal tissues | KYSE30, KYSE70, KYSE140, KYSE150, KYSE180, KYSE450, KYSE510, HET-1A | – | – | Have an important role in tumor cell proliferation and may be useful in diagnostic/therapeutic approaches | – | C.-M. Wang et al. (2014) |
| SPRY4-IT1 | 92 Patients with ESCC | KYSE-450, KYSE-510, KYSE-150, KYSE-180, KYSE-30, KYSE-70s, KYSE-140, Het-1A, Eca109 | – | – | Having a role in proliferation, migration, and invasion of tumor cells, it might be useful as a prognostic/therapeutic tool for patients with ESCC | Poor prognosis | H.-W. Xie et al. (2014) |
| HNF1A-AS1 | 25 Patients with EAC | HEEpiC, SKGT-4, OE33, FLO-1, JH-EsoAd1 | – | – | Can affect tumorigenesis of EAC cells | – | Yang et al. (2014) |
| FTH1P3 | 45 Paired of ESCC tissues and adjacent nontumorous samples | HET-1A, EC9706, EC1 | – | Sp1/NF-κB pathway | Having a vital role in tumorigenesis, it can be used as a therapeutic target for patients with ESCC | – | Ghaffar et al. (2018) |
| PEG10 | 43 Paired of EC samples and adjacent normal tissues | EC9706, KYSE150 | – | – | By improving the proliferation and invasion of tumor cells, it may act as a therapeutic target for patients with EC | – | Zang et al. (2015) |
- Abbreviations: DFS, disease-free survival; EAC, esophageal adenocarcinoma; EC, esophageal cancer; EMT, epithelial–mesenchymal transition; ESCC, esophageal squamous cell carcinoma; JAK2, Janus kinase 2; lncRNA, long noncoding RNA; NF-κB, nuclear factor-κB; OS, overall survival; STAT3, signal transducer and activator of transcription 3; TGF-β, transforming growth factor-β.
| lncRNA | Numbers of clinical samples | Assessed cell line | Signaling pathways | Function | Patient's prognosis | Reference |
|---|---|---|---|---|---|---|
| MEG3 | 28 Paired of patients with ESCC | EC109 | ER stress pathway | Can inhibit cell growth and induce apoptosis | Poor prognosis | Z.-L. Huang et al. (2017) |
| RP11-766N7.4 | 50 Patients with ESCC | TE-13, TE-1, EC-1, Eca-109 | EMT pathway | By acting as a tumor suppressor lncRNA, it may have some therapeutic/prognostic advantages | Correlated with poor OS | G.-L. Yao et al. (2017) |
| LOC285194 | 142 Tumor samples and paired adjacent normal tissues | KYSE30, KYSE 70, KYSE 150, KYSE510 Eca109, Het-1A | – | As a molecular marker may have importance in predicting patients’ prognosis after surgery | Low DFS and OS | Tong et al. (2014) |
- Abbreviations: DFS, disease-free survival; EMT, epithelial–mesenchymal transition; ER, endoplasmic reticulum; ESCC, esophageal squamous cell carcinoma; lncRNA, long noncoding RNA; OS, overall survival.
Among the oncogenic lncRNAs in esophageal cancer is SNHG16. This lncRNA acts as a “sponge” for miR-140-5p, thus modulating the expression of its target gene ZEB1 and enhancing epithelial–mesenchymal transition (EMT; K. Zhang, Chen, Song, & Chen, 2018). Globally, several oncogenic lncRNAs exert their role in esophageal cancer through similar mechanisms (Table 1).
Tumor suppressor lncRNAs have been shown to be downregulated in esophageal cancer cells. The epigenetic mechanism might be involved in this process. For instance, a comprehensive study of several cancer types including esophageal cancer has shown silencing of the lncRNA MORT in diverse malignancies by DNA methylation. Their observation led to the recognition of this lncRNA as one of the most frequently altered genes by epigenetic mechanisms in human cancers (Vrba & Futscher, 2018).
Although several lncRNAs have been shown to be dysregulated in both EAC and ESCC, some lncRNAs show subtype-specific expression patterns (W. Liu et al., 2018), thus can be used as diagnostic markers for differentiation of subtypes.
3 ASSOCIATIONS BETWEEN LncRNAs AND CANCER-RELATED SIGNALING PATHWAYS IN ESOPHAGEAL CANCER CELLS
The lncRNA CASC9 has been shown to induce the focal adhesion kinase-PI3K/AKT signaling pathways through cooperating with the cAMP response element-binding protein and enhancing expression of LAMC2 (Liang, Chen et al., 2018). TUG1 is associated with chemotherapy resistance in ESCC (L. Jiang et al., 2016). This lncRNA has a role in the regulation of PIK3/AKT and mitogen-activated protein kinase (MAPK) signaling pathways in lung cancer cells through modulation of HOXB7expression (E. Zhang et al., 2014). However, this effect has not been assessed in esophageal cancer. Linc01014 modulates gefitinib resistance in ESCC through regulation of estimated glomerular filtration rate (EGFR)–PI3K–AKT–mammalian target of rapamycin (mTOR) pathway (X. Fu, Cui, Liu, & Zhao, 2019). In addition, LINC01617 enhances the proliferation and metastasis of esophageal cancer cells through the AKT pathway (W. Liu et al., 2018). LINC01503 has a role in decreasing ERK2 dephosphorylation by DUSP6. Thus, LINC01503 overexpression results in the activation of extracellular signal-regulated kinase (ERK) signaling via MAPK. Moreover, this lncRNA interrupts the interaction between EBP1 and the p85 subunit of PI3K, thus enhancing AKT signaling (J.-J. Xie et al., 2018). On the other hand, the tumor suppressor lncRNA GAS5 inhibits ESCC proliferation and migration through suppression of PI3K/AKT/mTOR pathway (G. Wang, Sun, Zhao, & Li, 2018). Another tumor suppressor lncRNA, namely DESC1 has a proapoptotic effect in ESCC through downregulation of the EGFR/AKT pathway (Ng et al., 2016). Figure 2 shows a number of lncRNAs that are involved in the pathogenesis of esophageal cancer through modulation of the AKT pathway. The lncRNA SNHG1 has a prominent role in the EMT phenomenon through downregulating E-cadherin and upregulating vimentin and N-cadherin. This lncRNA also activates the Notch signaling pathway through increasing the Notch1 and Hes-1 expression levels (Y. Zhang et al., 2018). MALAT1 have a similar effect on EMT through enhancing Ezh2–Notch1 signaling pathway (Chen, Xia, Chen, Hu, & Yuan, 2018). Oncogenic roles of CCAT2 in esophageal cancer have been shown to exert through modulation of the Wnt signaling pathway (Wang & Wang, 2019). HERES, as a highly upregulated lncRNA in ESCC, instantaneously controls canonical and noncanonical Wnt signaling pathways through interrelation with EZH2 (You et al., 2019). On the other hand, the lncRNA UCA1 has been shown to suppress ESCC growth through altering the function of the Wnt signaling pathway (X. Wang et al., 2016). The lncRNA FTH1P3 increases the invasive and metastatic potential of ESCC through modulation of the SP1/nuclear factor-κB pathway (Ghaffar et al., 2018).
4 DIAGNOSTIC/PROGNOSTIC VALUE OF LncRNAs IN ESOPHAGEAL CANCER
In a previous study, Hao et al. (2015) have shown the ability of global coding and lncRNA signature in the differentiation of ESCC samples from neighboring noncancerous tissues. They also introduced the lncRNA ESCCAL-1 as one of the most dysregulated lncRNAs in ESCC samples (Hao et al., 2015). Wei et al. (2015) have implemented a comprehensive assessment of a custom-made combined lncRNA–mRNA microarray and RNA-sequencing (RNA-seq) data of ESCC samples and the corresponding noncancerous samples. They found several differentially expressed lncRNAs between these two sets of samples, the bulk of them exhibiting restricted expression signature. Moreover, the majority of these lncRNAs were associated with metastasis-related signaling pathways (Wei et al., 2015).
RP5-1172N10.2, RP11-89N17.4, LA16c-325D7.2, RP11-579D7.4, RP1-251M9.2, RP11-259O2.2, and LINC00173) which could predict survival of patients with ESCC. Functional enrichment analysis revealed that these lncRNAs might participate in the development of ESCC through modulation of cell cycle regulation, histone methylation, and cancer-related signaling pathways including PI3K/AKT and HIF-1 pathway (Mao et al., 2018). A recent analysis of RNA-seq data of patients with esophageal cancer provided from The Cancer Genome Atlas database has shown an association between expression levels of RPL34-AS1 and GK3P lncRNAs and overall survival of patients in an independent manner from clinical features. In silico enrichment analyses implied possible participation of these lncRNAs in the development of esophageal tumors (Miao, Sui, Zhang, & Yin, 2018). J. Yu et al. (2019) have used a bioinformatics method to identify lncRNA biomarkers in ESCC. They have recognized differential expression (DE) of 259 lncRNAs between early and advanced stage samples. Additional evaluations led to the identification of nine lncRNAs (AC098973, AL133493, RP11-51M24, RP11-317N8, RP11-834C11, RP11-69C17, LINC00471, LINC01193, and RP1-124C) whose expression profiles could predict tumor behavior and patients’ outcome. These lncRNAs mainly participated in cell cycle control and DNA replication (J. Yu et al., 2019). You et al. (2019) have identified six lncRNAs whose expression profiles were correlated with patients’ outcome. Two of these lncRNAs namely HERES and RP11-1L12.3 were signified as those increasing hazard ratios, while RP11-114H23.1, RP11-114H23.2, CTD-2319I12.1, and LINC00330 had the opposite effect. On the basis of the expression profile of these lncRNAs, esophageal cancer samples could be classified into four distinctive classes that correlate with patients’ survival or smoking history (You et al., 2019). H. Liu et al. (2019) have applied an in silico method to identify dysregulated lncRNA–miRNA–mRNA network in esophageal cancer. Dysregulated genes were enriched in chromosome segregation, extracellular matrix assembly, cell cycle, apoptosis, and cyclic GMP–protein kinase G signaling pathways. They also reported the correlation between expression of WDFY3-AS2, CASC8, UGDH-AS1, RAP2C-AS1, AC007128.1, AC016205.1, AC092803.2, and AC079949.2 lncRNAs clinical outcome (H. Liu et al., 2019). Another comprehensive analysis of lncRNAs profiles has led to the identification of seven upregulated lncRNAs and 21 downregulated lncRNAs whose signature defined poor overall survival of esophageal cancer patients, and 42 upregulated and 16 downregulated lncRNAs whose expression profiles were indicative of poor recurrence-free survival (W. Liu et al., 2018). Wang et al. (2017) have reported upregulation of HOTAIR in serum samples of patients with ESCC compared to normal controls. Serum levels of this lncRNA could differentiate patients from healthy controls with a diagnostic power of 0.793. In addition, expression levels of this lncRNA were correlated with tumor node metastasis stage (Wang et al., 2017). Table 3 summarizes the results of studies that reported the prognostic value of lncRNAs in esophageal cancer.
| Sample number | Kaplan–Meier analysis | Univariate Cox regression | Multivariate cox regression | Reference |
|---|---|---|---|---|
| 78 Pairs of esophageal cancer tissues and normal adjacent tissues | Patients with the high expression of DUXAP8 had a more unfavorable prognosis than those with low expression | – | – | Xu et al. (2018) |
| 96 Patients with ESCC | Patients with high levels of NEAT1 expression had shorter OS | NEAT1 expression, tumor size, lymph node metastasis, distant metastasis, and TNM stage were correlated with survival | NEAT1 expression was an independent risk factor for OS | Chen et al. (2015) |
| 100 Patients with ESCC and 56 adjacent nonneoplastic controls | Patients with high levels of HOTAIR expression had a shorter survival | – | In patients with ESCC, HOTAIR expression was correlated with the histological differentiation, tumor-nodule-metastasis classification, clinical stage, lymph node metastasis | X. Li et al. (2013) |
| 65 Patients with ESCC | Patients with a higher linc00460 expression had poorer overall survival | – | – | Liang et al. (2017) |
| 78 ESCC specimens | Patients with high levels of HOTAIR expression had a shorter survival time | – | In patients with ESCC, HOTAIR expression level, TNM stage, and lymph node metastasis were determined to be independent prognostic indicators for the OS rate | F. J. Chen et al. (2013) |
| 142 Tumor samples and paired adjacent normal tissues | Patients with low expression of LOC285194 had poorer OS and DFS | low expression of LOC285194 was correlated with CRT response | Low expression of LOC285194 and distant metastasis were independent prognosis factors that could affect the OS and DFS in patients with ESCC after esophagectomy | Tong et al. (2014) |
| 106 Patients with ESCC | Patients with high expression of NORAD had poorer OS and DFS | High NORAD expression was a risk factor for OS in patients with ESCC | NORAD expression and the UICC stage were independent prognostic factors in ESCC | X. Wu et al. (2017) |
| 137 ESCC tissues and adjacent normal samples | Patients with high levels of HOTAIR expression had a shorter survival | – | HOTAIR expression was an independent prognostic indicator for metastasis and death | Ge et al. (2013) |
| 132 Patients with ES | Patients with high MALAT1 expression had a shorter survival time | – | The expression of MALAT1 was an independent predictor for the survival of EC patients | C. Huang et al. (2016) |
| 82 ESCC tissues and adjacent noncancerous samples | Patients with high expression of FOXCUT had a worse prognosis | – | – | F. Pan et al. (2014) |
| 90 ESCC samples and adjacent nontumor tissues | Patients with high UCA1 expression had a poorer prognosis | UCA1 expression, clinical stage, tumor differentiation, and lymph node metastasis were correlated with the OS rate | UCA1 expression level, tumor differentiation, clinical stage, and lymph node metastasis were independent prognostic indicators for the OS | J.-Y. Li et al. (2014) |
| 87 Paired with ESCC samples | Patients with high ZEB1-AS1 expression had a poorer OS and DFS | – | In patients with ESCC after esophagectomy, tumor grade, ZEB1-AS1 expression, depth of invasion, and lymph node metastasis were independent prognostic factors for OS | Y.-L. Wang et al. (2015) |
| 218 Paired of ESCC and adjacent normal tissues | Patients with high TUG1 expression had a poor prognosis, especially for cases with well and moderate differentiation, chemotherapy-sensitive tumors, smaller size, and ulcerative type | LNM, TNM stage, and high TUG1 expression were related to poor prognosis | In patients with ESCC, TUG1 expression, LNM, and TNM stage were independent predictors of poor survival | L. Jiang et al. (2016) |
| 93 ESCC samples and adjacent normal tissues | Patients with high levels of HOTAIR had lower OS | High expression level of HOTAIR was correlated with clinical stage, TNM classification, histological differentiation, and vital status | HOTAIR expression was an independent factor for OS | F. J. Chen et al. (2013) |
| 142 Patients with ESCC and adjacent normal tissues | Patients with high expression of BANCR had poorer DFS and OS | – | In patients with ESCC after esophagectomy, high expression of BANCR was an independent predictor of poor survival | Z. Liu et al. (2016) |
| 106 Paired of ESCC tissues and adjacent noncancerous samples | Patients with MALAT1 high-expression level had a poorer OS | – | – | W. Wang et al. (2016) |
| 104 Patients with ESCC | Patients with PVT1 high-expression level had lower overall survival time | PVT1 expression level, differentiation status, N stage, and TNM stage were identified as prognostic factors | PVT1 expression was correlated with overall survival | X. Wu et al. (2017) |
| 66 Patients with ESCC and adjacent normal tissues | Patients with higher UCA1 expression had a poorer prognosis | – | – | Jiao et al. (2016) |
| 130 Cancerous tissues and adjacent noncancerous patients | Patients with higher PCAT-1 expression had a poorer survival time | – | High PCAT-1 expression and lymph node metastasis were an independent predictor of poor survival in ESCC | W.-H. Shi et al. (2015) |
| 92 Patients with ESCC | Patients with high levels of SPRY4-IT1 expression had lower overall survival time | – | SPRY4-IT1 expression, lymph node metastasis, and TNM stage were independent prognostic factors for the OS in patients with ESCC | H.-W. Xie et al. (2014) |
| 40 Paired of tumor and adjacent tissues | Patients with high HOTAIR expression had a poorer overall survival | – | – | Xu et al. (2017) |
| 50 Patients with ESCC | Patients in the high-expression group had a better overall survival rate | – | – | G.-L. Yao et al. (2017) |
- Abbreviations: DFS, disease-free survival; ESCC, esophageal squamous cell carcinoma; lncRNA, long noncoding RNA; LNM, lymph node metastasis; NF-κB, nuclear factor-κB; OS, overall survival; STAT3, signal transducer and activator of transcription 3; TGF-β, transforming growth factor-β; TNM, tumor node metastasis; UICC, Union for International Cancer Control.
5 GENOMIC VARIANTS WITHIN LncRNAs AND RISK OF ESOPHAGEAL CANCER
Copy number variations (CNVs) have been detected in genomic loci of lncRNAs in esophageal cancer samples. Recurrent gain CNV regions have been demonstrated in PVT1, LINC00887, and LINC00964, whereas LINC01415 and WEE2-AS1 were frequently lost in esophageal cancer (W. Liu et al., 2018). Moreover, a number of single-nucleotide polymorphisms (SNPs) have been identified in genomic regions that correspond to lncRNAs. Some of these SNPs have been associated with the risk of esophageal cancer. For instance, the rs920778 in the intronic enhancer region of the HOTAIR has been associated with risk of ESCC (X. Zhang et al., 2014). Consistent with this observation, T allele of this SNP increases expression of the oncogenic lncRNA HOTAIR (X. Zhang et al., 2014). The rs11752942 within lincRNA-uc003opf.1 exon has also been associated with risk of ESCC. The GG and AG genotypes of this SNP significantly decreased the risk of ESCC compared to the AA genotype. The underlying mechanism for this observation is the effect of the G allele in reducing expression of lincRNA-UC003opf.1 through targeting miRNA-149 (H. Wu et al., 2013).
6 APPLICATION OF LncRNAs AS THERAPEUTIC TARGETS IN ESOPHAGEAL CANCER
Aberrant expressions of lncRNAs in esophageal tumor specimens have been associated with resistance to chemoradiotherapy. X. L. Zhou et al. (2016) have assessed expression profile of lncRNAs in ESCC cell lines and their corresponding cisplatin-resistant cells and reported DE of AFAP1-AS1, UCA1, and HOTAIR between these two sets of cell lines. Validation of these results in pretreatment biopsy samples showed a correlation between high expression level of AFAP1-AS1 and poor response to definitive chemoradiotherapy (X. L. Zhou et al., 2016). Conversely, low expression of LOC285194 has been indicative of chemoresistance/radioresistance in patients with ESCC (Tong et al., 2014). LncRNA TUSC7 also modulates chemotherapy resistance of ESCC through regulation of miR-224 and DESC1 (Chang et al., 2018). Moreover, the WISP1-mediated radioresistance of ESCC has been shown to be correlated with upregulation of the lncRNA BOKAS (H. Zhang et al., 2015). The lncRNA FAM201A confers radioresistant phenotype in ESCC cells through modulation of expression of ATM and mTOR via sponging miR-101 (Chen, Liu et al., 2018). Moreover, TUG1 enhances cisplatin resistance in ESCC cells by modulating the expression of Nrf2 (Z. Zhang, Xiong, Li, Xu, & Guo, 2019).
The data presented above imply that targeted therapies against certain lncRNAs might enhance response of patients to conventional anticancer therapies. A number of in vivo studies have assessed the effects of suppression of oncogenic lncRNAs on suppression of tumor growth in animal models. For instance, Y. Xu et al. (2019) have evaluated antitumor effects of PVT1-specific antisense oligonucleotide (ASO) in a xenograft mice model of EAC. They also used specific ASO against YAP1, a gene which acts in a positive feedback circuit with PVT1. Notably, they demonstrated that simultaneous inhibition of these two genes significantly decreased tumor growth in vivo (Y. Xu et al., 2019). Chen, Liu et al. (2018) have confirmed the efficacy of FAM201A silencing in suppression of tumor growth and enhancement of radiosensitivity in the xenograft animal model. Moreover, in an animal model, K. Zhang et al. (2018) have shown slower growth of tumor cells that were transfected with short hairpin RNA against SNHG16 compared to controls.
7 EXPRESSION PROFILE OF MiRNAs IN ESOPHAGEAL CANCER
Several studies have reported up-/downregulation of miRNAs in esophageal cancer samples. On the basis of the direction of dysregulation, miRNAs can be divided to tumor suppressor miRNAs (Table 4) and onco-miRNAs (Table 5). miRNA signature not only discriminate normal tissues from tumoral tissues, it also distinguishes esophageal tumor histologies and recognize Barrett's esophagus patients who are at high risk for development of EAC (Feber et al., 2008).
| MicroRNA | Numbers of clinical samples | Assessed cell line | Targets/regulators | Signaling pathways | Function | Patient's prognosis | Reference |
|---|---|---|---|---|---|---|---|
| miR-381 | 3 Pairs of primary ESCC and recurrent ESCC in situ after radiotherapy | TE1, ECA109, EC9706, KYSE30, KYSE150, KYSE450 | CTNNB1, LEF1, CDK1, XIAP, CXCR4 | – | Overexpression of miR-381 could sensitize ESCC cells to irradiation and also could inhibit cell proliferation, migration, and invasion | – | S. Zhou et al. (2015) |
| miR-133a | 48 Pairs of ESCC tissues and adjacent normal tissues | KYSE-150, KYSE-510, EC-9706, SHEE, TE13 | Sox4, EMT | – | miR-133a could act as a tumor suppressor in ESCC through targeting Sox4 and the EMT process | – | S. Li et al. (2015) |
| miR-133a | Paraffin-embedded tissue blocks from 140 patients and 84 matched normal and tumor specimens | TE2, T.Tn | FSCN1, MMP14 | – | miR-133a could directly regulate FSCN1 and MMP14, hence it can serve as a strong tumor suppressor of ESCC | Poor prognosis | Akanuma et al. (2014) |
| miR-133a, miR-133b | 100 Pairs of ESCC tissues and adjacent normal tissues | – | – | – | The combined expression of miR-133a and miR-133b may predict chemosensitivity of patients with ESCC undergoing paclitaxel-based chemotherapy | Poor prognosis | G. Chen et al. (2014) |
| miR-133b | 47 Pairs of ESCC tissues and adjacent normal tissues | TE-1, TE-8, KYSE150, KYSE450, Eca-109, EC9706, HEEC, Het-1A | CUL4B | AKT/GSK3β/β-catenin | MiRNA-133b could inhibit cell proliferation and promote apoptosis in ESCC by targeting CUL4B | – | H. Huang et al. (2018) |
| miR-34a | 50 Pairs of ESCC tissues and adjacent normal tissues | – | – | – | miR-34a expression may be involved in the pathogenesis of EC | – | Asadi, Shanehbandi, Mohammadpour, Hashemzadeh, and Sepehri (2019) |
| miR-365 | 30 Pairs of ESCC tissues and adjacent normal tissues | EC109, EC1, EC9706, HEEC, HEK293T | PSAT1 | – | Overexpression of miR-365 could inhibit colony formation, cell invasion, and growth in esophageal cancer cell lines in vitro, and also tumor development in vivo, by modulating expression levels of PSAT1 | – | Qiao et al. (2018) |
| miR-340 | 64 Pairs of ESCC tissues and adjacent normal tissues | HEEC, HEK293T, EC1, EC109, EC9706 | PSAT1 | – | Overexpression of miR-340 could inhibit ESCC cell growth, invasion, colony formation, and tumor growth in a xenograft mouse model | – | Yan et al. (2015) |
| miR-506 | 28 Pairs of ESCC tissues and adjacent normal tissues | Eca109, Kyse30, TE-1, NEEC | CREB1 | – | Overexpression of miR-506 could suppress the proliferation of esophageal cancer cells by targeting CREB1 | – | W.-J. Yao et al. (2015) |
| miR-197 | 46 Pairs of ESCC tissues and adjacent normal tissues | – | – | – | miR-197 expression was downregulated in EC with poor prognosis, therefore, it could be a biomarker to predict outcome and treatment response | Poor prognosis | T. Wang et al. (2014) |
| miR-218 | Serum samples of 106 patients with EC and 60 healthy volunteers | – | – | – | The serum expression of miR-218, tumor differentiation, stage, and lymph node metastasis are closely related; therefore, it is suggested that miR-218 is a tumor suppressor | Poor prognosis | Z. Jiang et al. (2015) |
| miR-873 | 36 Pairs of EC tissues and adjacent normal tissues | EC-109, EC-1, TE-1, TE-10, KYSE-150, HEEC | DEC2 | – | miR-873 could modulate EC cell growth, migration, and invasion by directly suppressing DEC2 | Poor prognosis | Liang, Zhang et al. (2018) |
| miR-145 | 40 Pairs of ESCC tissues and adjacent normal tissues | Ec109, EC9706, TE-1, TE-10, KYSE150, KYSE140 | ERK5 | – | miR-143 could act as a tumor suppressor in ESCC by targeting ERK-5 | – | Y. Ni et al. (2013) |
| miR-27a | 30 Pairs of ESCC tissues and adjacent normal tissues | TE-1, TE-10, TE-11, ECA-109, HEEC, Het-1A | KRAS | – | miR-27a could inhibit cell proliferation in ESCC by targeting KRAS | – | Y. Jiang, Duan, and Zhou (2015) |
| miR-375 | 10 Pairs of ESCC tissues and adjacent normal tissues | EC109, Het-1A | MTDH | – | miR-375 could suppress EC cell growth and invasion by repressing MTDH | – | Hu et al. (2017) |
| miR-377 | 30 Pairs of ESCC tissues and adjacent normal tissues, serum samples of serum of 114 patients with ESCC and 50 healthy individuals | KYSE30, KYSE70, KYSE150, KYSE270, KYSE410, 45 HKESC-146, T.Tn47, HEK293 | CD133, VEGF | – | miR-377 could suppress the initiation and progression of EC via inhibiting CD133 and VEGF | Poor prognosis | Feng, Bai, Yu, Wang, and Liu (2017) |
| miR-486 | 10 Pairs of ESCC tissues and adjacent normal tissues | KYSE150, EC9706, TE-9, Het-1A | CDK4, BCAS2 | – | miR-486 could act as a tumor suppressor by targeting CDK4 and BCAS2 and affect cell proliferation, colony formation, and apoptosis | – | Lang and Zhao (2018) |
| miR-99a, miR-100 | 101 Pairs of ESCC tissues and adjacent normal tissues | KYSE-450, EC9706, | mTOR | – | miR-99a/100 could induce tumor cell apoptosis by targeting mTOR | Poor prognosis | Sun et al. (2013) |
| miR-625 | 158 Pairs of EC tissues and adjacent normal tissues | Eca-109, TE-1 | Sox2 | – | Downregulation of miR-625 could promote proliferation and invasion in EC by targeting Sox2 | – | Z. Wang et al. (2014) |
| miR-155 | 26 Pairs of EC tissues and adjacent normal tissues | ECA109, KYSE150, EC18, EC1, ECA-9706, HEEC, Het-1A | FGF2 | – | miR-155 could upregulate TNF-α, IL-12, and iNOS, while downregulating IL-10, Arg-1, and IL-22 levels in the culture medium from tumor-associated macrophages (TAMs) Overexpression of miR-155 in TAMs could suppress cell viability, migration, and invasion of ECA109 cells and also could reduce angiogenesis | – | P. Wang, Xu, Qin, Zhang, and Zhuang (2018) |
| miR-153 | 28 Pairs of ESCC tissues and adjacent normal tissues | OE-21, HET1A | SNAI1 | – | miR-153 could inhibit tumor progression in ESCC via targeting SNAI. Suppression of miR-153 could dictate SNAI1 upregulation during EMT and metastatic progression of ESCC | – | Zuo et al. (2016) |
| miR-382 | 46 patients with ESCC including poor outcome group (n = 28) and good outcome group (n = 18) | – | – | – | miR-382 could be considered as a potential predictive biomarker for both outcome prognosis and treatment of ESCC | Poor prognosis | Qi et al. (2015) |
| miR-203 | 32 Pairs of ESCC tissues and adjacent normal tissues | EC9706, KYSE150 | BMI-1 | – | miR-203 was downregulated in side population cells MiR-203 could inhibit the proliferation and self-renewal of EC stem-like cells by suppressing BMI-1 | – | X. Yu et al. (2013) |
- Abbreviations: AKT, protein kinase B; EC, esophageal cancer; EMT, epithelial–mesenchymal transition; ESCC, esophageal squamous cell carcinoma; IL-10, interleukin-10; IL-12, interleukin-12; IL-22, interleukin-22; iNOS, inducible nitric oxide synthase; miRNA, microRNA; mTOR, mammalian target of rapamycin; TNF-α, tumor necrosis factor-α; VEGH, vascular endothelial growth factor.
| MicroRNA | Numbers of clinical samples | Assessed cell line | Targets/regulators | Signaling pathways | Function | Patient's prognosis | Reference |
|---|---|---|---|---|---|---|---|
| miR-330–3p | 35 Pairs of ESCC tissues and adjacent normal tissues | EC109, KYSE150, HET-1A | PDCD4 | – | Overexpression of miR-330–3p could promote ESCC cell proliferation, survival, and migration. It could decrease G1 phase cells and increase S/G2/M phase in EC109 cells | – | Meng et al. (2015) |
| miR-212 | 46 Pairs of ESCC tissues and adjacent normal tissues | – | – | – | Overexpression of miR-212 was observed in EC patients with poor prognoses, therefore it could be considered as a marker for predicting the outcome, treatment response, and treatment method for EC | Poor prognosis | Qi et al. (2014) |
| miR-548-3p, miR-576-5p | 52 Pairs of ESCC tissues and adjacent normal tissues | SHEE, SHEEC, EC109, KYSE150, KYSE450, KYSE30, TE3, 293T | NRIP1 | – | MiR-548-3p and miR-576-5p via downregulating NRIP1 could enhance the migration and invasion of ESCC cells | – | X. Ni et al. (2018) |
| miR-374a | 46 Pairs of EC tissues and adjacent normal tissues | Eca109, Kyse30, Kyse140, Kyse180, Kyse410, Kyse510, Kyse520, TE-1, NEEC | Axin2 | – | MiR-374a via suppressing Axin2 could promote EC cell proliferation | – | Wang, Xin et al. (2015) |
| miR-367 | Serum samples from 35 patients with ESCC and from 35 healthy control patients, 46 pairs of ESCC tissues, and adjacent normal tissues | Kyse30, TE-1 | – | – | Lentivirus-induced miR-367 downregulation could inhibit cancer growth and cell cycle transition in TE-1 and Kyse30 cells | Poor prognosis | Sun, Song, Feng, and Gao (2016) |
| miR-21 | 89 Pairs of EC tissues and adjacent normal tissues | TE11 | PTEN | PI3K/AKT | MiR-21 via targeting PTEN/PI3K/AKT pathway could inhibit apoptosis and promote proliferation, migration, invasion, and cell cycle | – | Y.-R. Wu, Qi, Deng, Luo, and Yang (2016) |
| miR-21 | 16 Pairs of ESCC tissues and adjacent normal tissues | EC9706, EC-1 | FASL, TIMP3, RECK | – | Downregulation of miR-21 via targeting FASL, TIMP3, and RECK could suppress cell growth, invasion, and also induce cell apoptosis | Poor prognosis | N. Xu et al. (2013) |
| miR-1322 | 44 Pairs of ESCC tissues and adjacent normal tissues, serum samples from 201 patients with ESCC and 201 healthy controls | EC9706, EC109, KYSE150, HEK293 | ECRG2 | – | MiR-1322 could downregulate the ECRG2 with the TCA3 allele and could not downregulate the ECRG2 with the TCA4 allele | – | T. Zhang et al. (2013) |
| miR-145 | GSE43732 datasets | TE8, TE11, OE33, FLO1 | SMAD5 | – | MiR-145 by targeting SMAD5 could promote EC cell proliferation and metastasis | Poor prognosis | Q. Zhang, Gan, Song, Chai, and Wu (2018) |
| miR-502 | 13 Pairs of ESCC tissues and adjacent normal tissues | TE-1 | – | AKT | MiR-502 by promoting AKT phosphorylation could mediate EC cell TE1 proliferation | – | J. Xu, Pan, and Hu (2018) |
| miR-1290 | 24 Pairs of ESCC tissues and adjacent normal tissues | Eca109, TE13 | SCAI | – | MiR-1290 by targeting the antioncogene SCAI could promote ESCC cell proliferation, migration, and invasion | – | M. Li et al. (2015) |
- Abbreviations: AKT, protein kinase B; EC, esophageal cancer; ESCC, esophageal squamous cell carcinoma; miRNA, microRNA; PI3K, phosphoinositide 3-kinase; PTEN, phosphatase and tensin homolog.
8 DIAGNOSTIC AND PROGNOSTIC ROLE OF MiRNAs IN ESOPHAGEAL CANCER
On the basis of availability of miRNAs in body fluids and their association with course of esophageal cancer, miRNAs are potential diagnostic and prognostic markers. A systematic review and meta-analysis of literature have shown that expression levels of miR-21, miR-133a, miR-133b, miR-138, miR-203, miR-375, and miR-655 in tissue samples and expression levels of miR-21 and miR-223 in blood could be regarded as prognostic biomarkers for esophageal cancer (Gao, Zhao, Zhang, Zhang, & Wu, 2018). Diagnostic potential of circulating miRNAs has also been assessed in another meta-analysis which showed overexpression of miR-21 and miR-223 and downregulation of miR-375 in patients with ESCC compared to healthy controls. The area under the curve values were 0.80, 0.73, and 0.69 for miR-21, miR-223, and miR-375, respectively. These values were higher in discrimination of patients with early ESCC and the noninvasive carcinoma stage from controls (L. Zhang et al., 2018). Table 6 summarizes the studies which investigated biomarker role of miRNAs in esophageal cancer.
| Sample number/study design | Area under curve | Sensitivity | Specificity | Kaplan–Meier analysis | Univariate Cox regression | Multivariate cox regression | Reference |
|---|---|---|---|---|---|---|---|
| 84 Matched normal and tumor specimens | – | – | – | There was a significant decrease in the OS in the low miR-133a group | – | Positive MMP14 staining, positive FSCN1 staining, the N stage, and the T stage were identified as significant independent prognostic factors | Akanuma et al. (2014) |
| 100 Pairs of ESCC tissues and adjacent normal tissues | – | – | – | The OS of ESCC patients with lower expression of miR-133a/miR-133b was shorter than the higher expression of them | Tumor stage and combined expression of miR-133a/b were independent prognosis factors in patients with ESCC | The expression of miR-133a/b, tumor stage, and tumor differentiation were independent prognostic factors for OS in patients with ESCC | G. Chen et al. (2014) |
| 46 Pairs of ESCC tissues and adjacent normal tissues | – | – | – | MiR-197 expression level was correlated with survival time, and patients with lower expression had shorter OS | – | Tumor length and miR-197 expression were correlated with survival time | T. Wang et al. (2014) |
| 46 Pairs of ESCC tissues and adjacent normal tissues | – | – | – | Expression of miR-212 was correlated with survival time and patients with higher miR-212 expression had longer survival times | – | Tumor size and miR-212 expression were correlated with survival time | Qi et al. (2014) |
| Serum levels of miRNA-218 in 106 patients with EC and 60 healthy volunteers | 0.833 | 71.7% | 76.7% | – | – | – | Z. Jiang et al. (2015) |
| 101 Pairs of ESCC tissues and adjacent normal tissues | – | – | – | Low expression of miR-99a/100 was associated with poor OS in patients with ESCC | TNM stage was also associated with OS | miR-99a/100 expression and TNM stage were unfavorable prognostic factors | Sun et al. (2013) |
| Serum samples from 35 patients with ESCC and from 35 healthy control patients, 46 pairs of ESCC tissues and adjacent normal tissues | OS was shorter in patients with high serum miR-367 level | Serum miR-367, TNM stage, differentiation grade, and lymph node metastasis were closely correlated with the survival of patients with ESCC | Lymph node metastasis and serum miR-367 were independent factors to predict survival in patients with ESCC | Sun et al. (2016) | |||
| 46 Patients with ESCC including poor outcome group (n = 28) and good outcome group (n = 18) | – | – | – | miR-382 expression had a significant reverse correlation with ESS patient survival time | – | Cox single factor related risk analysis showed that the miR-382 level, tumor size, TNM stage, postoperative time, and patient survival time had significant correlations | Qi et al. (2015) |
| Assessment of expression of miRNA-1322 in 44 pairs of ESCC tissues and adjacent normal tissues, serum samples from 201 patients with ESCC, and 201 healthy controls | 0.847 | 81.7% | 82.5% | – | – | – | T. Zhang et al. (2013) |
| GSE43732 datasets | – | – | – | Poor prognosis was associated with high expression of miR-145 | – | – | Q. Zhang et al. (2018) |
- Abbreviations: ESCC, esophageal squamous cell carcinoma; miRNA, microRNA; OS, overall survival; TNM, tumor node metastasis.
9 ASSOCIATION BETWEEN EXPRESSION PROFILE OF MiRNAs AND RESISTANCE TO ANTICANCER MODALITIES
Aberrant expression of miRNAs in esophageal cancer samples has been associated with resistance of these cells to therapeutic modalities. Hummel et al. (2014) have constructed an in vitro model of secondary chemotherapy resistance in EAC and ESCC cells. They compared the miRNA signature between cisplatin or 5-fluorouracil (5-FU) resistant variants and chemotherapy-sensitive controls. Notably, they detected specific miRNA profiles in cisplatin and 5-FU resistant cells which were also different from EAC and ESCC cells. Among the most dysregulated miRNAs were miR-27b-3p, miR-193b-3p, miR-192-5p, miR-378a-3p, miR-125a-5p, and miR-18a-3p which are involved in the regulation of expression of chemotherapy response-related genes KRAS, TYMS, ABCC3, CBL-B, and ERBB2 (Hummel et al., 2014). Ma et al. (2016) have demonstrated the association between miRNA-196a and expression of ABCG2, an ATP-binding cassette protein that is involved in pumping the cisplatin molecules out of the cell. This finding potentiates this miRNA as a molecule that is involved in resistance to cisplatin. Wang, Feng et al. (2015) showed the role of miRNA-499 in the modulation of the activity of DNA polymerase-β and enhancement of the response to cisplatin. The role of miRNA-223 in inhibition of PARP1 and induction of apoptosis has potentiated this miRNA as a possible determinant of response to chemotherapeutic agents (Streppel et al., 2013). Downregulation of miRNA-200c has also been associated with response to cisplatin therapy in esophageal cancer (Tanaka et al., 2013). Other miRNAs that regulate cell apoptosis might be involved in the modulation of response to chemotherapeutic agents (Vrana et al., 2017). The sensitivity of esophageal cancer cells to radiotherapy is also altered by the expression profile of certain miRNAs. For instance, miRNA-31 suppresses the expression of a number of DNA repair enzymes, thus enhancing the radiosensitivity of esophageal cancer cells (Lynam-Lennon et al., 2012). miRNA-22 modulates radioresistance possibly through modulation of expression of RAD51, a DNA repair enzyme that participates in the homologous recombination (X.-C. Wang et al., 2012). Besides, miRNA-98 increases radioresistance by interacting with BCL2 (Jin et al., 2016).
10 DISCUSSION
While the worldwide incidence of ESCC is decreasing due to the economic enhancement and nutritional rectification (Thun et al., 2017), statistics show a rapid increase in the incidence rates of EAC in some regions (Arnold, Laversanne, Brown, Devesa, & Bray, 2017). Although modification of lifestyles might slow the trend, identification of molecular mechanisms for the pathogenesis of esophageal cancer is an important necessity for controlling this cancer. Expression profiling and mechanistical studies have shown that lncRNAs exert fundamental roles in the pathogenesis of esophageal cancer. Although these studies have led to the identification of downstream targets of several lncRNAs, for other lncRNAs, this field needs to be explored in the future. Among pathways regulated by lncRNAs in esophageal cancer are PI3K/AKT and HIF-1 pathways (Mao et al., 2018). These interconnected pathways have essential roles in the pathogenesis of esophageal cancer (Zeng et al., 2016). Notably, drugs targeting the PI3K/AKT have been shown to inhibit ESCC in preclinical studies (N. Shi, Yu, & Chen, 2019) and are being tested in clinical trials (Sadeghi & Gerber, 2012). Thus, a future perspective of this study area is the identification of lncRNAs that modulate the response of patients to these kinds of targeted therapies.
While expression profiling of lncRNAs has its own beneficial effects in the understanding, the pathophysiology of esophageal cancer, integrative assessment of lncRNA and mRNA profiles, and construction of interactive network have more practical application in identification of novel key regulators in this type of cancer (Alaei, Sadeghi, Najafi, & Masoudi-Nejad, 2019). Availability of combined lncRNA–mRNA microarray and RNA-seq datasets of esophageal cancer tissues and corresponding noncancerous tissues has speeded up recognition of these interactive networks. A prototype of these methods is the reannotation of expression profile, DE analysis, and assessment of coexpressed lncRNAs and protein-coding genes (PCG). This methodology has led to the construction of lncRNAs–PCGs networks and subtype-specific modules that could differentiate ESCC, EAC, and Barrett's esophagus (Z. Liu et al., 2016). Moreover, these kinds of studies would help us in recognition of the underlying causes of differences in the clinical outcomes of patients who were in the same stage and received similar therapeutic options. Such differences have been reported in clinical settings and are mostly attributed to the heterogeneity of tumors at the molecular level (Navin et al., 2011). Furthermore, clarification of the functional roles of the lncRNA–miRNA–mRNA networks in the pathogenesis of esophageal cancer would offer innovative beneficial targets for therapeutic interventions.
Taken together, lncRNAs and miRNAs are regarded as putative biomarkers in esophageal cancer whose expression profiles not only predict tumor behavior but also can differentiate subtypes/stages of esophageal cancer. They also can be used as therapeutic targets in esophageal cancer, if the efficacy of lncRNA/miRNA-targeted therapies is approved in the preclinical studies. The DE of miRNAs/lncRNAs between precancerous conditions and esophageal cancer would facilitate the recognition of biomarkers for cancer progression and identification of molecular mechanisms that are involved in this process. The advent of biomarkers for early detection of cancer along with the design of targeted therapies based on the data obtained from mechanistical studies would improve survival of patients with esophageal cancer.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
M. T. and S. G.-F. supervised the study, wrote the draft, and edited the submission. H. S, W. B., and S. D. performed the data collection.
Open Research
DATA AVAILABILITY STATEMENT
The analyzed datasets generated during the study are available from the corresponding author on reasonable request.