Highly upregulated in liver cancer (HULC): An update on its role in carcinogenesis
Abstract
Highly upregulated in liver cancer (HULC) was initially recognized during the screening of a hepatocellular carcinoma (HCC)-specific gene library. Further studies demonstrated its aberrant upregulation in several other tumor types. The oncogenic roles of this long noncoding RNA (lncRNA) have been verified through expression studies as well as functional studies. Moreover, the results of knockdown experiments have indicated diminished carcinogenic effects of cancer cell line in nude mice following HULC silencing. More recent studies have shown that expression levels of this lncRNA might be used as diagnostic biomarkers in cancer patients. Moreover, mechanistical studies have revealed associations between HULC and two HCC-related viruses namely hepatitis B and C viruses. Taken together, HULC can be regarded as a therapeutic target not only for HCC but also for a variety of human malignancies. In the current review, we summarized the recent literature about the role of HULC in the carcinogenesis and its potential application in cancer diagnosis and prognosis.
1 INTRODUCTION
Highly upregulated in liver cancer (HULC) was initially recognized during the screening of a hepatocellular carcinoma (HCC)-specific gene library as the utmost upregulated long noncoding RNA (lncRNA) in this type of cancer (Panzitt et al., 2007). Being located on chromosome 6p24.3, HULC spans a region of approximately 1,600 nucleotides and has two exons (Panzitt et al., 2007). The transcribed RNA does not have considerable open-reading frame and does not produce any protein. HULC promoter and its first exon are located in a long-terminal repeat (LTR) retrotransposon-like sequence (Kapusta et al., 2013). Subsequent studies revealed overexpression of this lncRNA in a variety of cancers including gastric (Y. Zhao et al., 2014), pancreatic (Peng, Gao, & Feng, 2014), and osteosarcoma (Sun, Yang, Geng, Wang, & Zhang, 2015). The fundamental roles of this lncRNA in the tumorigenesis process have been certified through both in vitro (Y. Zhao et al., 2014) and in vivo studies (Cui et al., 2015; Z. Lu et al., 2016).
Several environmental and internal factors regulate transcription of this lncRNA. Among the most appreciated environmental factors is hepatitis B virus (HBV; Matouk et al., 2009). Moreover, cAMP response element-binding protein (CREB) can upregulate expression of HULC by binding with a region on proximal site of HULC promoter. Phospho-CREB can preserve an open configuration across the HULC promoter. HULC has an inhibitory effect on several miRNAs including miR-372. Suppression of miR-372 diminishes translational repression of Prkacb, which can enhance CREB phosphorylation (J. Wang et al., 2010). Cholesterol can enhance HULC transcription through retinoid X receptor α (RXRA), a transcription factor, which is involved in the pathogenesis of HCC cells (Cui et al., 2015). Expression of HULC is also regulated by another lncRNA, namely cancer-upregulated drug-resistant (CUDR), which is upregulated in several cancers. This effect is exerted through suppression of HULC promoter methylation and is paralleled with malignant transformation of embryonic stem-cell-originated hepatocyte-like cells (Gui, Li, Li, Pu, & Lu, 2015).
Expression of HULC can also be regulated by posttranscriptional mechanisms. HULC binding with IGF2 mRNA-binding protein 1 (IGFBP1) reduces the half-life and constant expression of this lncRNA. IGF2BP1 might enhance HULC degradation by increasing HULC deadenylation (Hammerle et al., 2013). Besides, miR-203 has a role in posttranscriptional regulation of HULC (Wan et al., 2016). Based on the presence of diverse regulatory mechanisms for fine-tuning of HULC expression, dysregulation of its expression in cancers might be due to disruption of several mechanisms. In this review, we provided data regarding the role of HULC in carcinogenesis and the putative underlying mechanisms. We also summarized the available evidence, which points to the role of this lncRNA as a diagnostic/prognostic marker.
2 MECHANISM OF HULC INVOLVEMENT IN CARCINOGENESIS
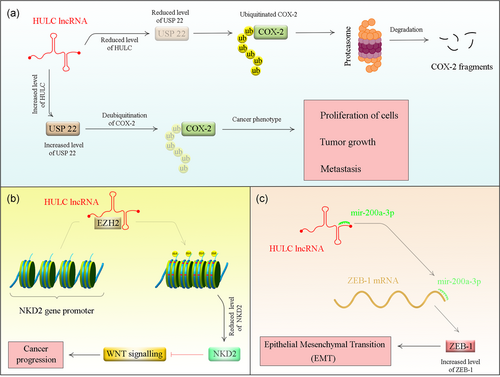
Consistent with the prominent role of HULC in HCC pathogenesis, most of the mechanistical studies have been conducted in HCC. For instance, Wang et al. have shown positive correlations between expression levels of HULC and the oncogene high mobility group A2 (HMGA2). They also reported the role of HULC in upregulation of HMGA2 in HCC cells. Their mechanistical studies revealed that HULC acts as a sponge miR-186 thus abrogating the miR-186-mediated HMGA2 suppression. Notably, HMGA2 silencing diminished the HULC-mediated growth of HCC cells both in cell lines and in animal models (Y. Wang, Chen, et al., 2017).
HULC might also affect immune responses. This effect has been verified in HBV-related liver cirrhosis where HULC has been shown to enhance differentiation of regulatory T cells through direct downregulation of p18 (J. Zhao et al., 2015). Downregulation of p18 by HULC has also been shown to enhance hepatoma cell proliferation (Du et al., 2012).
In addition, HULC collaborates with another oncogenic lncRNA, namely metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) to enhance liver cancer stem cells' growth. Wu et al. have demonstrated higher levels of these two lncRNAs and telomere repeat-binding factor 2 (TRF2) in human HCC tissues. Upregulation of both HULC and MALAT1 increased recruitment of RNA pol II, P300, and CREPT on the promoter of TRF2 leading to the overexpression of TRF2. TRF2 interacts with the mentioned lncRNAs to construct a complex on the telomeric region, which protects telomere and enhances its elongation (Wu et al., 2016).
HULC also enhances cancer-related phenotypes such as cell proliferation, migration, and invasion and suppresses cisplatin-mediated apoptosis. This lncRNA specifically binds to Y-box binding protein 1 (YB-1) protein, an essential constituent of translationally inactive messenger ribonucleoprotein elements, which preserves mRNA in an inactive form. HULC binding with YB-1 enhances its phosphorylation resulting in the dissociation of YB-1 from its restrained mRNA. Such dissociation leads to translation of silenced oncogenic mRNAs such as cyclin D1, cyclin E1, and matrix metalloproteinase 3 (D. Li et al., 2017).
Figure 1 shows a summary of proposed mechanisms for participation of HULC in the pathogenesis of cancer.
3 ABNORMAL EXPRESSION OF HULC IN CANCERS
Abnormal upregulation of HULC has been demonstrated in a wide range of human malignancies. Table 1 shows the results of studies that assessed expression of this lncRNA in cancer cell lines. These results indicate that HULC has a prominent role in induction of invasiveness in these cell lines.
| Cancer type | Targets/regulators and signaling pathways | Assessed cell lines | Function | Reference |
|---|---|---|---|---|
| Gastric cancer | – | GES-1, MGC-803, MKN-45 | ∆ HULC: ↓ apoptosis, ↑ sensitivity to chemotherapeutic drugs | Y. Zhang, Song, Wang, Hu, and Jiang (2016) |
| – | GES-1, SGC7901, MKN28, MKN45, AGS, BGC823 | ∆ HULC: ↓invasion, ↓migration | Y. Zhao et al. (2014) | |
| Non-small-cell lung cancer | SPHK1/PI3K-Akt | NCI-H23, NCI-H522 | ↑ HULC: ↑cell proliferation, ↓apoptosis | L. Liu et al. (2018) |
| PI3K-Akt | A549, SPC-A1, H23, NCI-H520, 16HBE | ∆HULC: ↓cell proliferation, ↓cell migration, ↓invasion | J. Zhang, Lu, Zhu, and Yang (2016) | |
| Prostate cancer | – | LNCaP, PC3, DU145, RWPE-1 | ∆HULC: ↓cell proliferation, ↓invasion | Zheng et al. (2018) |
| Epithelial ovarian carcinoma | ATG7 and ITGB1 | OVCAR3, A2780 | ↓HULC: ↓cell proliferation, ↓migration, ↓invasion | S. Chen, Wu, et al. (2017) |
| Cholangiocarcinoma | CXCR4/miR-372 and miR-373 | QBC939, SK-cha-1, RBE, HEK293T | ↑ HULC: ↑migration, ↑invasion | W.-T. Wang et al. (2016) |
| Hepatocellular carcinoma | miR15a and PTEN/AKT-PI3K-mTOR | Hep3B | ↑ HULC: ↑cancer cell growth, ↑colony formation | Xin et al. (2018) |
| HMGA2 | HepG2, Huh7, HepG2.2.15, 293T | ↑ HULC: ↑proliferation | Y. Wang et al. (2017) | |
| miR-107/E2F1/SPHK1 signaling | HepG2, Huh7, HepG2.2.15, 293T | ↑ HULC: ↑tumor angiogenesis | Z. Lu et al. (2016) | |
| miR-200a-3p/ZEB1 | Huh-6, Huh-7,HepG2, BEL-7402, MHCC-97H, Sk-Hep1, SMMC-7721, L02 | ∆HULC: ↓proliferation, ↑apoptosis | S. P. Li et al. (2016) | |
| COX-2 protein | HepG2, Hep3B | ↑ HULC: ↑tumor growth | Xiong et al. (2017) | |
| Colon cancer | RTKN | HIEC, FHC, NCM460, GEO, SNU-C1,Colo205, HCT-116, HT-29, SW480 | ∆HULC: ↓proliferation, ↓colony formation | Dong, Wei, Lu, and Bi (2019) |
| Osteosarcoma | – | MG-63, U2OS, SAOS-2, hFOB | ∆HULC: ↓proliferation, ↓migration, ↓invasion | Sun et al. (2015) |
| miR-122/PI3K-AKT, JAK-STAT and Notch pathways | MG63, hFOB1.19 | ∆HULC: ↓cell viability, ↓migration, ↓invasion, ↑apoptosis | Kong and Wang (2018) | |
| Nasopharyngeal carcinoma | p53-p21 | SUNE, CNE-1 | ∆HULC: ↑cell cycle arrest, ↑cell apoptosis | Jiang and Liu (2017) |
| Chronic myeloid leukemia | c-Myc/PI3K-Akt | K562, KG-1, THP-1 | ∆HULC: ↓proliferation, ↑apoptosis | Y. Lu et al. (2017) |
| Glioma | ESM-1/PI3K-Akt-mTOR | U87MG, U251 | ∆HULC: ↓proliferation, ↓angiogenesis, ↓invasion, ↓migration | Guan et al. (2016) |
| Diffuse large B-cell lymphoma (DLBCL) | – | OCI–LY–3, OCI–LY–7, OCI–LY–10,SU–DHL–4, SU–DHL–6, RCK–8 | ∆HULC: ↓tumor growth, ↑apoptosis | Peng, Wu, and Feng (2016) |
| Pancreatic cancer | – | MIAPace-2, CFPAC-1, PANC-1, AsPC-1, SW1990, BxPC-3 | ∆HULC: ↓proliferation, ↓tumor growth | Peng et al. (2014) |
| Colorectal carcinoma | NKD2 | DLD1, HCT116, LOVO, RKO, LS174T, HCT8, HR28348, HT29, SW620, SW480, NCM460 | ∆HULC: ↓proliferation, ↓migration, ↓invasion, ↑apoptosis | Yang et al. (2016) |
| Bladder cancer | ZIC2/PI3K-AKT | T24, RT4 | ∆HULC: ↓proliferation, ↑apoptosis | J. Wang, Ma, and Liu (2017) |
A number of studies have validated the results of cell line studies in animal models. As summarized in Table 2, HULC silencing results in decreased tumorigenic potential, while upregulation of this lncRNA increases tumor formation, tumor size, and tumorigenesis.
| Cancer type | Animal models | Function and comments | Reference |
|---|---|---|---|
| Prostate cancer | Nude mice | ∆ HULC: ↓cell growth | Zheng et al. (2018) |
| Epithelial ovarian carcinoma | Female BALB/c nude mice | ↑HULC: ↑tumorigenesis | S. Chen, Wu, et al. (2017) |
| Hepatocellular carcinoma | Balb/C mice | ↑HULC: ↑tumor weight, ↑tumor formation rate, ↓differentiation | Xin et al. (2018) |
| Nude mice | ↑HULC: ↑tumor weight, ↑tumor volume | Wang et al. (2017) |
Finally, overexpression of HULC has been reported in several cancer types (Table 3). Such overexpression was mostly correlated with poor patients' outcome.
| Cancer type | Numbers of clinical samples (tissues, serum, and so forth) | Kaplan–Meier analysis | Univariate cox regression | Multivariate cox regression | Reference |
|---|---|---|---|---|---|
| Gastric cancer | 42 Gastric cancer (GC) tissues, 42 GC plasma and 25 normal control | High HULC expression was significantly associated with poor survival time | – | – | J. Zhang, Lu, et al. (2016) |
| 173 GC patient serums (30 patients with gastric polyps, 30 patients with high atypical hyperplasia or intestinal metaplasia) and 110 age-matched normal controls | High HULC expression was significantly associated with poor OS | – | – | Jin et al. (2016) | |
| Cervical cancer | 244 Paired primary cervical cancer tissues and adjacent normal tissues | High HULC expression was associated with poorer outcome | HULC was identified as a prognostic factor for OS | HULC was identified as an independent and significant prognostic factor for 5-year survival rates | Wang et al. (2016) |
| Non-small-cell lung cancer | 102 Patients with NSCLC | High HULC expression was correlated with low OS | – | – | L. Liu et al. (2018) |
| 58 NSCLC patients | High HULC expression was correlated with shorter OS time | – | – | J. Zhang, Lu, et al. (2016) | |
| Hepatocellular carcinoma | 30 HCC patients and 20 healthy control | – | – | – | Xie, Ma, and Zhou (2013) |
| 30 Liver cancer tissues | – | – | – | Xin et al. (2018) | |
| 38 HCC patients and 21 normal controls | High HULC expression was significantly associated with a poor 5-year OS rate | – | – | Yang et al. (2016) | |
| Prostate cancer | 53 PCa tissues and adjacent nontumor tissues | High HULC expression was associated with poor OS | – | – | Zheng et al. (2018) |
| Epithelial ovarian carcinoma | 96 Cancerous tissues and 15 normal samples | – | – | – | S. Chen, Wu, et al. (2017) |
| Colon cancer | 67 Paired colon cancer tissues and adjacent nontumor tissues | High HULC expression was significantly correlated with worse OS | – | – | Dong et al. (2019) |
| Nasopharyngeal carcinoma | 22 Paired NPC and adjacent normal tissues | High HULC expression was related to poorer outcome | – | High expression of HULC was an independent prognostic factor for OS of NPC patients and not affected by TNM classification and clinical stage | Jiang and Liu (2017) |
| Glioma | 70 Glioma patients | High HULC can predict poor OS | High HULC levels were significantly associated with OS | Remained as an independent predictor for OS | Yan et al. (2017) |
| Pancreatic cancer | 304 Tumor and adjacent normal tissues | High HULC expression was significantly associated with shorter survival time | HULC expression, tumor size, lymph node metastasis, and vascular invasion were prognostic indicators | HULC expression was an independent prognostic indicator for OS | Peng et al. (2014) |
| Diffuse large B-cell lymphoma (DLBCL) | 142 DLBCL patients and 60 samples with reactive lymph nodes as controls | High HULC expression showed remarkably shorter OS and progression-free survival (PFS) | HULC expression was an independent factor that forecasts OS and PFS | HULC expression was a significant independent predictor of poor survival of DLBCL patients | Peng et al. (2016) |
| Bladder cancer | 276 Bladder cancer tissues | High HULC expression was related to lower recurrence-free rate | – | – | S. Chen, Wu, et al. (2017) |
| Colorectal carcinoma | 35 pairs of CRC and adjacent nontumor tissues | High HULC expression was related to a poorer OS | – | – | Yang, Huang, Peng, Hou, and Liu (2016) |
| Osteosarcoma | 78 Osteosarcoma patients | High HULC expression was significantly associated with OS rate | HULC expression was significantly related to clinical stage and distant metastasis, which can affect the overall survival of osteosarcoma patients | HULC was independent prognostic factor with clinical stage and distant metastasis | Sun et al. (2015) |
| 33 Patient samples | HULC expression level was not significantly associated with clinicopathological features of patients | – | – | Uzan et al. (2016) |
A previous meta-analysis of 10 studies has shown that HULC transcript level serves as an independent prognostic biomarker for poor overall survival and metastasis in human cancers (X. Chen, Lun, et al., 2017). The studies included pancreatic cancer, osteosarcoma, gastric cancer, large B-cell lymphoma, and cervical cancer (X. Chen, Lun, et al., 2017). Further studies in other cancer types also validate their results (summarized in Table 3).
4 ASSOCIATIONS BETWEEN THE EXPRESSION OF HULC AND VIRAL INFECTIONS
Considering the obvious overexpression of HULC in HCC and the causal link between HCC and HBV, researchers have assessed the associations between HULC overexpression and EBV infection. The first evidence for such association has been obtained from Matouk et al. (2009) study, which reported overexpression of HULC in two HCC cell lines producing HBV compared with their original lines that lack HBV. This study indicated a possible role for HBV in upregulation of HULC. Further studies showed that positive correlations between expression levels of HULC and expression of HBV X protein (HBx) in clinical samples of HCC. Furthermore, authors have shown the role of this HBV protein in upregulation of HULC in both human immortalized normal liver cells and hepatoma cells (Du et al., 2012). Another study has shown concurrent upregulation of HULC and hepatitis B X-interacting protein (HBXIP) in HBV-infected patients, especially those with HBV-associated HCC. Moreover, RNA immunoprecipitation assay has shown interaction between HULC and HBXIP (Ruan et al., 2018). A recent study has indicated upregulation of HULC during hepatitis C (HCV) infection and its role in changing the lipid pool to facilitate virus life cycle. Consistent with the role of HULC in lipid biosynthesis, this lncRNA had a critical role in enhancement of the quantities of lipid droplets in infected cells. Notably, HULC silencing has abrogated the association between HCV-core protein and lipid driblets. Moreover, upregulation of HULC in HCV-infected cells was mediated by RXRA, a process which was facilitated by HCV-core protein (Sharma, Tripathi, & Das, 2019).
5 HULC VARIANTS AND RISK OF CANCER
Hapmap database has shown high linkage disequilibrium (LD) between HULC single nucleotide polymorphisms (SNPs) (Huang, Zhang, & Shao, 2018). For instance, the most assessed SNP within this gene (rs7763881) is in complete LD with the other SNP located in the promoter of gene (rs1328867). The wild-type allele of rs1328867 (T allele) is anticipated to bind with some transcription factors such as C-Myc (Y. Liu et al., 2012). Based on the high level of LD between HULC SNPs, most studies have genotyped only the rs7763881. Liu et al. have genotyped the rs7763881 in HULC in 1300 HBV-positive HCC patients, 1344 HBV-chronic carriers, and 1344 individuals with HBV clearance. Authors have reported decreased HCC risk for carriers of AC and CC genotypes of rs7763881 compared with AA genotype carriers. Yet, there was no significant association between rs7763881 and HBV clearance. Consequently, authors deduced a role of the rs7763881 in decreasing the risk of HCC in HBV-persistent carriers (Y. Liu et al., 2012). Shaker et al. have assessed the association between rs7763881 and risk of colorectal cancer (CRC) and adenomatous polyps (AP) in120 CRC patients, 30 AP patients, and 96 healthy individuals. The AC genotype of the rs7763881 has shown to exert protective effects against CRC. Patients who had AA genotype of rs7763881 had higher serum HULC levels compared with those that had AC genotype (Shaker, Senousy, & Elbaz, 2017). Kang et al. (2015) have genotyped the rs7763881 A/C SNPs in 380 esophageal squamous cell carcinoma (ESCC) cases and 380 normal individuals and reported a possible association between this SNP and decreased risk of cancer. A recent meta-analysis of three studies in cancer patients (CRC, HCC, and esophageal squamous cell carcinoma) has demonstrated association between the rs7763881 A/C polymorphism and lower risk of cancer in allelic, heterozygote, and dominant models compared with the wild-type AA genotype (Huang et al., 2018).
6 DIAGNOSTIC VALUE OF HULC
Diagnostic value of HULC has been assessed in a number of cancers and cancer-related situations. For instance, a previous study in Egyptian population has demonstrated higher diagnostic value of HULC in differentiation of CRC from healthy state compared with its value in differentiation of AP from healthy state (Shaker et al., 2017). Table 4 summarizes the results of studies that addressed the diagnostic value of HULC in human cancers.
| Cancer type | Numbers of clinical samples | Distinguish between | Area Under Curve | Sensitivity | Specificity | Reference |
|---|---|---|---|---|---|---|
| Non-small cell lung cancer (NSCLC) | 102 Patients with NSCLC | High HULC group versus low HULC group | 0.9392 | – | – | L. Liu et al. (2018) |
| Hepatocellular carcinoma (HCC) | 30 HCC patients and 20 healthy control | Tumor versus control samples | 0.86 | – | – | Xie et al. (2013) |
| Osteosarcoma | 33 OS tissues | Good outcome patients versus poorer outcome patients | 0.48 | 50.0 | 73.7 | Uzan et al. (2016) |
| Diffuse large B-cell lymphoma (DLBCL) | 142 DLBCL patients and 60 samples with reactive lymph nodes as controls | Cancerous versus normal samples | 0.97 | – | – | Peng et al. (2016) |
| Gastric cancer (GC) | 173 GC patient serums, 30 patients with gastric polyps, 30 patients with high atypical hyperplasia or intestinal metaplasia) and 110 age-matched normal controls | GC patients and healthy controls | 0.88 | 82.0 | 83.6 | Jin et al. (2016) |
| 58 Samples of GC tissues and adjacent non-cancer tissues | GC tissues and non-tumorous tissues | 0.769 | 70.7 | 72.4 | Y. Zhao et al. (2014) | |
| Pancreatic cancer | 304 Tumor and adjacent normal tissues | Tumor and normal samples | 0.977 | – | – | Peng et al. (2014) |
7 HULC EXPRESSION IN PERIPHERAL BLOOD OF CANCER PATIENTS
Jin et al. have assessed the expression of HULC in serum samples of 100 primary gastric cancer patients, 30 polyp patients, and 110 normal individuals. They reported high levels of this lncRNA in cancer patients. Notably, they demonstrated correlations between high serum HULC markers of aggressive behavior of tumor (Jin et al., 2016). Xian et al. have reported higher levels of HULC in the plasma of preoperative gastric cancer patients compared with the plasma of gastrointestinal stromal tumor (GIST) patients, gastritis/peptic ulcer patients, and healthy individuals. Such observation indicated the significance of this lncRNA in diagnosis of gastric cancer (Xian, Zhuo, Sun, Liang, & Zhao, 2018).
8 DISCUSSION
Several studies have shown upregulation of HULC in human cancers. Consistent with these results, certain SNPs within this gene have shown to influence the expression of transcript and modulate risk of cancer. So, HULC can be regarded as a risk locus in several cancers and in different population. The results of functional studies have indicated a role for this lncRNA in posttranscriptional regulation of a number of tumor suppressor miRNAs, so the most possible mechanism for participation of this lncRNA in tumorigenesis is its function as a competing endogenous RNA. Diagnostic power of this lncRNA in some cancer types such as diffuse large B-cell lymphoma, hepatocellular carcinoma, and lung, gastric, and pancreatic cancers has been excellent. Notably, the availability of blood as a suitable source for expression analysis of this lncRNA facilitates follow-up of patients and early detection of cancer recurrence. It is expected that expression analysis of this lncRNA in body fluids substitute more aggressive methods for follow-up of cancer patients. Although knockdown studies have shown a decrease in tumorigenic potential of cancer cells in animal models, the results of such studies are not sufficient to propose this strategy for human subjects. Consequently, it is necessary to validate these results in other animal models and with other cancer cell types to pave the way for design of novel anticancer modalities. A distinct feature of this lncRNA is its association with viral proteins and its role in the pathogenesis of virus-related cancers. This feature potentiates HULC-targeted therapies for these kinds of cancers.
Taken together, consistent results from cell line experiments, functional studies in animal models and human studies have shown HULC as an oncogenic lncRNA and a potential diagnostic and prognostic marker for malignancy.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
M. T. and S. G. F. supervised the study, wrote the draft, and edited the submission. M. H. E. and M. S. performed the data collection.
Open Research
DATA AVAILABILITY STATEMENT
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.