Long noncoding RNA Gm6135 functions as a competitive endogenous RNA to regulate toll-like receptor 4 expression by sponging miR-203-3p in diabetic nephropathy
Abstract
We aim to explore the relationship between Gm6135 and diabetic nephropathy. We detected the relative expression levels of Gm6135 and toll-like receptor 4 (TLR4) in diabetic nephropathy mice and high-glucose-cultured mouse mesangial cells SV40-MES-13 by the quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and western blot detection. Cell proliferation and apoptosis were detected after small interfering RNA (siRNA) interference or plasmid overexpression of Gm6135/TLR4, and bioinformatics method was used to predict and screen miR-203 as an intermediate factor. Through dual-luciferase reporter gene, RNA pull-down, qRT-PCR, and western blot, the binding relationship between Gm6135, miR-203-3p, and TLR4 was confirmed. The possibility of the competing endogenous RNA mechanism was demonstrated by cell localization assays and rip assays. Finally, the proliferation of mouse mesangial cells SV40-MES-13 was detected after mimics and inhibitor of microRNA, which were reversed with TLR4 overexpression and siRNA. The results showed that the relative expression levels of Gm6135 and TLR4 in the kidney and high-glucose-cultured mouse mesangial cells of diabetic nephropathy mice increased significantly. Overexpression or downregulation of Gm6135/TLR4 significantly affected the proliferation and apoptosis of mouse mesangial cells. Gm6135 upregulates TLR4 by competitively binding to miR-203-3p.
1 INTRODUCTION
Diabetes is a chronic metabolic syndrome characterized by hyperglycemia. It is estimated that by 2030, the number of diabetic patients worldwide will exceed 430 million (Zare Sakhvidi, Zare Sakhvidi, Mehrparvar, Foraster, & Dadvand, 2018). Diabetic nephropathy (DN) is one of the most serious complications of diabetes. Approximately 40% of diabetic patients develop DN, which is the leading cause of end-stage renal disease (Chu et al., 2018). DN has become a major public health problem worldwide, with enormous social and economic burdens for individuals, families, and society. Therefore, the research on the molecular mechanism of the development of DN not only provide a new perspective on the pathogenesis of DN, but also provide a more targeted program for the prevention and treatment of DN.
Renal structural and functional damage during the development of DN is ultimately clinically characterized by abnormal urinary albumin (Alicic, Johnson, & Tuttle, 2018). In recent years, researchers have proposed a microinflammation theory for the progression of DN, which is considered to be a natural immunity and low-grade inflammatory disease (Ichinose, Kawasaki, & Eguchi, 2007). Studies have confirmed that in DN, toll-like receptor 4 (TLR4) tumor necrosis factor-α (TNF-α), interleukin-18 (IL-18), and other inflammatory factors exist abnormal transcriptional expression (Ichinose et al., 2007). Abnormal expression of TLR4 has also been shown to be closely related to the progression of DN in animal and clinical studies. However, the specific activation mechanism of TLR4 in DN is not clear.
Noncoding RNA (ncRNA) accounts for approximately 98% of gene transcription products, including microRNAs (miRNAs) and long noncoding RNAs (lncRNAs). MiRNAs have been shown to play an important role in the pathogenesis of a variety of neoplastic diseases (Rak, Marczewska, & Wlodarski, 2016). LncRNAs are a class of ncRNA molecules greater than 200 nucleotides in length that are widely present in the nucleus and cytoplasm and are not involved or rarely involved in protein coding (Carpenter, 2016). However, lncRNA can play a role in various biological processes, such as cell proliferation and apoptosis, through various molecular mechanisms (Zhou et al., 2016). For example, J. Chen et al., 2018 proposed that the long noncoding RNA LnRPT is regulated by PDGF-BB and modulates the proliferation of pulmonary artery smooth muscle cells. LncRNAs can participate in gene expression regulation at transcriptional and posttranscriptional levels (Y. Li et al., 2018). In recent years, many studies have suggested that lncRNA can function as a competing endogenous RNA (ceRNA) (H. Li et al., 2017; Xie et al., 2017). CeRNA is also known as miRNA “molecular sponge,” which means the lncRNA can play a biological role by “absorbing” miRNAs and affecting the binding of miRNAs to downstream target genes.
By analyzing the results of mouse DN microassay, we found that Gm6135 was highly expressed in DN mice (S. Chen et al., 2017). Subsequently, we compared the gene expression levels of kidney tissue in mice with type 2 DN and normal mice by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and found that Gm6135 did show abnormally high expression in the kidney of DN mice. Subsequently, we predicted the target gene of Gm6135 by using starbase, RegRNA, and other software and found that miR-203 is its potential binding target. By reading the literature, it was found that miR-203 participates in many tumor formation by affecting cell proliferation and other functions (Chi et al., 2017; Lin et al., 2016; Liu et al., 2017). The occurrence of DN is closely related to the excessive proliferation of mesangial cells (Han et al., 2017; Wang et al., 2018). Therefore, we want to further explore whether Gm6135 will lead to excessive proliferation of mesangial cells through the expression of miR-203, and ultimately participate in the pathogenesis of DN.
2 MATERIALS AND METHODS
2.1 Experimental animal
Four-week-old specific pathogen free (SPF)-grade diabetic model mice (db/db mice) and control normal mice (db/m mice) were purchased from the Institute of Model Animals, Nanjing University. During the experiment, the mice were kept under constant temperature conditions, the standard feed and drinking water were free, and the light alternated for 12 hr. Tissue collection: The mice were killed after anesthesia, and the mice were quickly stripped and frozen in liquid nitrogen for later extraction of RNA and protein. All experimental animals were operated in accordance with the Qingpu Branch of Zhongshan Hospital Affiliated to Fudan University Ethics Committee standards.
2.2 Mouse biochemical index detection
The weight of the mice was weighed using a balance scale. The blood glucose concentration was measured using a fast blood glucose meter with the blood that was collected from the tail vein of the mouse. The 24-hr urine sample was collected by a mouse metabolic cage, meanwhile 1 ml of preservative was added to the container before collection.
2.3 Cell culture and transfection
Mouse mesangial cells SV40-MES-13 (ATCC, Manassas VA) were cultured in Dulbecco s modified Eagle medium (Hyclone, UT) containing 10% fetal bovine serum, 100 U/ml penicillin, and 100 U/ml streptomycin. After the cell fusion degree is approximately 80%, the 0.25% trypsin solution is digested, and the cell morphology changes from a spindle shape to a circular shape. The medium is added to terminate the digestion, and the cells are collected by centrifugation and passaged 1:3. Sugar treatment: After treating cells with 5 mM low sugar-mesangial cell (L-MC) and 25 mM high sugar-mesangial cell (H-MC) sugar concentration medium for 24 hr, the cells were harvested and RNA was collected to detect the expression of Gm6135/TLR4. After the cell fusion degree was approximately 60%, the serum-containing medium was removed and washed once with phosphate-buffered saline (PBS). After the PBS was removed, the transfection reagent mixed with Lipofectamine 3000 (Invitrogen, CA) was added to the culture dish, and a certain amount of serum-free medium was added and mixed for 20 min. After 6 hours, the serum-free medium was changed to serum-containing medium, and the transfection effect was verified by qRT-PCR. The overexpression plasmids and small interfering RNA (siRNAs) of miR-203 mimics or inhibitor and Gm6135/TLR4 used in this experiment were provided by Shanghai Jima Co., Ltd.
2.4 Quantitative reverse transcription-polymerase chain reaction
According to the instructions for rapid extraction of Trizol (Life Technologies, CA), total RNA was centrifuged at 4°C, the upper layer of water isopropanol precipitate was taken, washed, and dried at room temperature. Finally, 20–30 μl of diethyl pyrocarbonate treated water (DEPC) was added, and the RNA concentration was measured and stored in a −80°C refrigerator for use. Reverse transcription experiments were performed according to the instructions of the Takara OneStep PrimeScript MiRNA cDNA Synthesis Kit (Tokyo, Japan). PCR detection was carried out by the SYBR Green I fluorescent dye method, and the Gm6135 primer sequence was F: 5′-CATGGGATGTGAGCAGTCTT-3′, R: 5′-TGAGGATTCAGGCTGGAGTG-3′. The TLR4 primer sequence is F: 5′-ATGGCATGGCTTACACCACC-3′, R: 5′-GAGGCCAATTTTGTCTCCACA-3′. The relative concentration of the sample to be tested = ; the same experiment was repeated three times, and the average value was taken.
2.5 Dual-luciferase reporter gene
Plasmid construction: The binding sites of miR-203 and Gm6135/TLR4 were predicted by Target Scan. This binding sequence was sent to the company to construct wild-type and mutant plasmids. The mimics and inhibitors of miR-203 were subsequently ordered at Genepharma (Shanghai, China). The relative fluorescence values of the plasmids after transfection were then measured by a standardized method. The same experiment was repeated three times, and the average was taken.
2.6 Western blot detection
The total protein of mouse kidney tissue and SV40-MES-13 cells was extracted and quantified, and the protein concentration was adjusted for sodium dodecyl sulphate protein electrophoresis. After electrophoresis, it was transferred to a polyvinylidene fluoride (PVDF) membrane and routinely immunostained. The membrane was immersed in an antidilution solution and placed in a refrigerator at 4°C overnight, with a primary antibody (Invitrogen, Shanghai, China) dilution ratio of 1:500. The working concentration of the anti-rabbit secondary antibody (Beyotime, Nantong, China) was 1:1,000, and the membrane was incubated at 37°C for 2 hr, followed by chemiluminescence, development, fixation, and photographing. The same experiment was repeated three times and the average was taken.
2.7 Cell proliferation
SV40-MES-13 cells in logarithmic growth phase were selected and cultured in 96-well plates at a cell density of 1 × 106/ml, 100 μl per well. After 24 hr, 10 μL cell counting kit-8 (CCK8) was added to each well, and the light absorbance value was measured at 450 nm after 1 hr. Also, the 5-Ethynyl-2’-deoxyuridine (EDU) assay was taken to detect the proliferation of cells. All experiments were repeated three times independently.
2.8 Cell apoptosis detection
The cells were digested with trypsin containing no EDTA into the six wells and then resuspended by centrifugation in cold PBS. Subsequently, suspension cells were collected not less than 1 × 105/ml, and after centrifugation, 100 μl of 1× annexin buffer was added to suspend the cells. The specific method using annexin V–FITC labeling was as follows: 5 μl of annexin V and 1 μl of propidium iodide (PI) was stained, mixed, protected from light, and incubated at room temperature for about 15 min. After staining, 400 μl of 1× buffer was added to measure apoptosis on a flow cytometer. Three replicate wells were set in each group, and the experiment was repeated three times.
2.9 Cellular localization
The number of cells was grown to 1 × 106, and 200 μl of Lysis Buffer J was added to the flask to fully lyse the cells. After centrifugation, the supernatant contains cytoplasmic RNA and the remaining liquid contains nuclear RNA. After transferring the supernatant, buffer SK and absolute ethanol were added to the liquid containing cytoplasmic RNA and nuclear RNA, and cytoplasmic RNA and nuclear RNA were eluted by column centrifugation.
2.10 RNA binding protein immunoprecipitation
Follow the steps in the Millipore kit instructions. After the cells were lysed, the detection antibody was added, and the working concentration of the antibody was 8 μg per reaction system, and the mixture was incubated at 4°C overnight, and then rewarmed at room temperature for 1 hr. Protein G magnetic beads were used to capture the complex, and after washing, the RNA was extracted. After reverse transcription amplification, qRT-PCR detects RNA levels.
2.11 RNA pull-down assay
SV40-MES-13 cells were transfected with biotinylated miR-203-WT, miR-203-Mut, and negative control (GenePharma, Shanghai, China) respectively, and then harvested. Cell lysates were incubated with Dynabeads M-280 Streptavidin (Invitrogen, Shanghai, China) according to the manufacturer’s protocol. Similarly, the synthesized biotinylated DNA probe complementary to Gm6135 (GenePharma) was incubated with Dynabeads M-280 Streptavidin (Invitrogen) to generate probe-coated beads. Then, cell lysates were incubated with the probe-coated beads at 4°C overnight. The RNA complexes bound to these beads were eluted and detected by qRT-PCR analysis.
2.12 Data analysis
Data analysis was performed using SPSS 22.0 software and Graphpad software. Measurement data are expressed as mean ± standard deviation, Student’s t test was performed, and the difference was considered statistically significant when the p value was less than 0.05.
3 RESULTS
3.1 DN mouse biochemical indicators
We used normal db/m mice as controls to determine the progression of nephropathy in db/db diabetic mice. As shown in Figure 1a–c, we detect changes in body weight, blood glucose, and urinary albumin between db/m mice and db/db mice. It was found that the weight, blood glucose, and urinary albumin levels were significantly increased in the db/db mice at the eighth week.
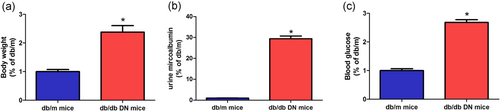
Biochemical indicators of diabetic nephropathy mice. After DN modeling, (a) db/db mice weight increased, (b) urine protein levels increased, and (c) blood glucose levels increased. The data are presented as mean ± standard deviation. *p < 0.05, according to Student’s t test. DN: diabetic nephropathy [Color figure can be viewed at wileyonlinelibrary.com]
3.2 Gm6135/TLR4 expression in DN
According to the literature, the expression of lncRNA in the kidneys of db/db mice and db/m mice was detected by lncRNA microassay (Chen et al., 2017), and it was found that Gm6135 was upregulated in the kidney of db/db mice. As shown in Figure 2a, Gm6135 was significant upregulation in the kidneys of 8-week-old db/db mice compared with normal db/m mice. TLR4 plays an important role in the occurrence of DN (Jheng et al., 2015; Yang, Zhang, Wang, Shi, & Zhao, 2017). The expression of TLR4 in db/db mice was also significantly increased (Figure 2b,c). To clarify the expression trend of Gm6135 in mesangial cells in a high glucose environment in vitro, different concentrations of glucose cultured mesangial cells were used in the experiment. As can be seen from Figure 2d, the relative expression of Gm6135 was significantly higher in the H-MC group compared with the L-MC group. The expression of TLR4 in SV40-MES-13 cells (H-MC group) cultured in high glucose (25 mmol/L) was also significantly increased (Figure 2e,f). The results indicate that high glucose concentration can stimulate the expression of Gm6135/TLR4 in SV40-MES-13 cells in a simulated diabetic environment in vitro.
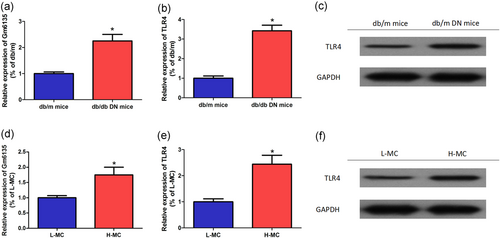
Gm6135/TLR4 expression in DN. (a) Compared with normal db/m mice, Gm6135 was significantly upregulated in db/db mouse kidneys. (b,c) TLR4 was upregulated in db/db mouse kidneys compared with normal db/m mice. (d) The relative expression of Gm6135 was significantly higher in the H-MC group compared with the L-MC group. (e,f) The expression of TLR4 in SV40-MES-13 cells (H-MC group) cultured in high glucose (25 mmol/L) was also significantly increased. The data are presented as mean ± standard deviation. *p < 0.05, according to Student’s t test. DN: diabetic nephropathy; H-MC: high sugar-mesangial cell; L-MC: low sugar-mesangial cell; TLR4: toll-like receptor 4 [Color figure can be viewed at wileyonlinelibrary.com]
3.3 Gm6135/TLR4 facilitates cell proliferation and inhibits cell apoptosis
To further investigate the role of Gm6135/TLR4 in the development of DN, we transfected SV40-MES-13 cells for 24 hr and then extracted RNA to detect the relative expression level of Gm6135/TLR4 by qRT-PCR. As shown in Figure 3a,b, the relative expression level of Gm6135/TLR4 was significantly lower in the H-MC siRNA group transfected with Gm6135/TLR4 siRNA than in the H-MC-NC group (p < 0.05); the relative expression level of Gm6135/TLR4 in the L-MC overexpression (OE) group with Gm6135/TLR4 overexpression plasmid was significantly higher than that in the L-MC-NC group (p < 0.05). Thus, expression of miR-203 in the SV40-MES-13 cell line can be downregulated or upregulated by transfection of Gm6135/TLR4 siRNA or overexpression plasmid. Through CCK8 and EDU experiments, we found that the proliferation of SV40-MES-13 cells was significantly decreased in the H-MC siRNA group transfected with Gm6135/TLR4 siRNA (p < 0.05); in the L-MC OE group, which was transfected with the Gm6135/TLR4 overexpression plasmid, the proliferative capacity of SV40-MES-13 cells was significantly increased (Figure 3c,d). Therefore, SV40-MES-13 cell proliferation ability can be inhibited or promoted by transfection of Gm6135/TLR4 siRNA or overexpression plasmid. We found that the apoptosis rate of SV40-MES-13 cells was significantly increased in the H-MC siRNA group transfected with Gm6135/TLR4 siRNA by the apoptosis assay (p < 0.05); Gm6135/TLR4 overexpression plasmid was transfected in the L-MC OE group, and the proportion of apoptosis in SV40-MES-13 cells was significantly decreased (p < 0.05), as shown in Figure 3e. Therefore, the proportion of apoptosis in SV40-MES-13 cells can be increased or decreased by transfection of Gm6135/TLR4 siRNA or overexpression plasmid.
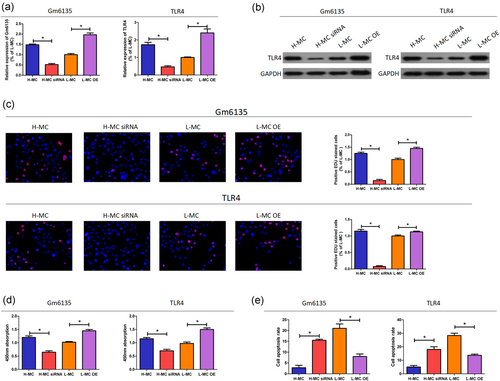
Gm6135/TLR4 facilitates cell proliferation and inhibits cell apoptosis. (a,b) Changes in Gm6135/TLR4 expression levels after interference and overexpression. (c,d) Changes in the proliferation of SV40-MES-13 cells after interference and overexpression. (e) Changes in apopotosis of SV40-MES-13 cells after interference and overexpression. The data are presented as mean ± standard deviation. *p < 0.05, according to Student’s t test. TLR4: toll-like receptor 4 [Color figure can be viewed at wileyonlinelibrary.com]
3.4 Interaction between Gm6135 and miR-203
A large number of studies have shown that lncRNA can regulate the expression of target genes by competitively binding miRNA as ceRNA (Li et al., 2017; Xie et al., 2017). Through binding site predictions, we found that miR-203 may bind to Gm6135 (Figure 4a). At the same time, miR-203 is highly homologous in humans and mice. To further verify the interaction between miR-203 and Gm6135, we performed a dual-luciferase reporter assay. Reporter gene results indicate that miR-203 can bind to Gm6135 (Figure 4b). The specific interaction between miR-203 and Gm6135 was further confirmed by the RNA pull-down assay using biotinylated miR-203. The binding sequence prediction of Gm6135 and miR-203 is shown in Figure 4c. RNA pull-down experiments showed that Gm6135 binds to miR-203 (Figure 4d). These results demonstrate that the recognition of Gm6135 by miR-203 is performed in a sequence-specific manner. We further investigated whether there is a negative correlation between Gm6135 and miR-203 in DN. First, overexpression of Gm6135 significantly reduced the level of miR-203, but the Gm6135 mutant lacking the miR-203 target site did not. (Figure 4e). Furthermore, miR-203 inhibitor enhanced Gm6135 expression and miR-203 mimics decreased Gm6135 expression compared with the negative control (NC) group (Figure 4f).
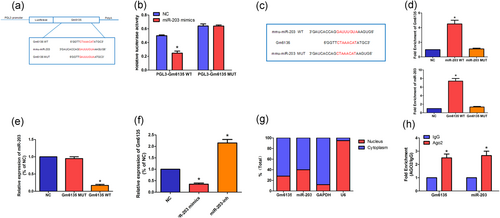
Interaction between Gm6135 and miR-203. (a) Gm6135 and miR-203 binding site prediction. (b) Reporter gene experiments show that Gm6135 can bind to miR-203. (c) MiR-203 mutant sequence construction. (d) RNA pull-down experiments show that miR-203 can bind to Gm6135, and (e) overexpression Gm6135 can inhibit the expression of miR-203. (f) Overexpression of miR-203 can inhibit the expression of Gm6135. (g) Cellular localization assays show that Gm6135 and miR-203 are mainly located in the cytoplasm. (h) RIP assays show that Gm6135 was regulated by miR-203 via an Ago2-induced RISC. The data are presented as mean ± standard deviation. *p < 0.05, according to Student’s t test. RIP: RNA binding protein immunoprecipitation; RISC: RNA-induced silencing complex [Color figure can be viewed at wileyonlinelibrary.com]
In fact, Gm6135 is primarily localized to the cytoplasm (Figure 4g), which means that Gm6135 can participate in posttranslational modification. Studies have shown that lncRNAs that are involved in posttranslational modification have potential as ceRNAs (Rashid, Shah, & Shan, 2016). Therefore, we intend to verify the possibility of Gm6135 as a ceRNA by rip experiment. The rip experiment showed that Gm6135 was regulated by miR-203 via an Ago2-induced RNA-induced silencing complex (RISC) (Figure. 4h). These results indicate that Gm6135 can function as an miR-203 ceRNA.
3.5 Gm6135 induces TLR4 via downregulation of miR-203
Through site prediction and crossover, we found that miR-203 also binds to TLR4 and its binding sequence is identical to the previous one (Figure 5a). Therefore, we hypothesized that Gm6135 can regulate the expression of TLR4 by miR-203. To address our hypothesis, we performed a dual-luciferase reporter gene analysis using the TLR4-WT and TLR4-MUT reporter genes. Our data showed a significant decrease in luciferase signal in cells cotransfected with miR-203 mimics and TLR4-WT compared with cells cotransfected with NC and TLR4-WT (Figure 5b). At the same time, we found that miR-203 inhibitor significantly increased TLR4 expression, whereas miR-203 mimics reduced TLR4 expression (Figure 5c), indicating that miR-203 recognizes TLR4 and destabilizes TLR4 mRNA. Since TLR4 mRNA is mainly located in the cytoplasm (Figure 5d), we expect that miR-203 can regulate TLR4 mRNA in the cytoplasm via the RNAi pathway. As expected, anti-Ago2 RIP showed a two- to threefold increase in total TLR4 mRNA and miR-203 enrichment compared with the nonspecific immunoglobulin G RIP. (Figure. 5e). We also found that miR-203 inhibitor abolished the downregulation of TLR4 induced by Gm6135 siRNA (Figure 5f,g). The above-mentioned results indicate that Gm6135 regulates the expression of TLR4 by binding to miR-203.
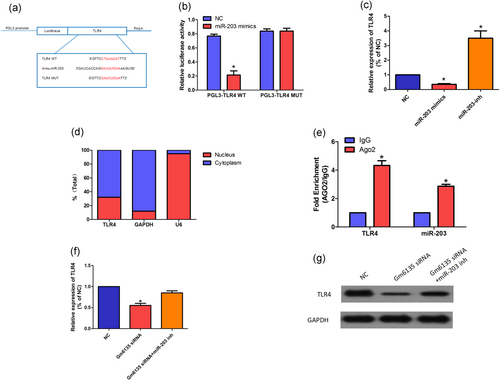
Gm6135 induces TLR4 via downregulation of miR-203. (a) Putative binding sequences between miR-203 and TLR4-WT/TLR4-MUT. (b) Luciferase activities in SV40-MES-13 cells cotransfected with miR-203 mimics or NC and luciferase reporters containing TLR4-WT or TLR4-MUT are shown. (c) Relative expression of TLR4 in SV40-MES-13 cells treated with miR-203 mimics or inhibitor. (d) Subcellular location of TLR4 in SV40-MES-13 cells. (e) Relative enrichment of miR-203 and TLR4 in anti-Ago2 pull-down using indicated SV40-MES-13 cells. TLR4 expression was normalized to β-actin levels and miR-203 expression was normalized to U6 small RNA expression. (f,g) The relative level of TLR4 in SV40-MES-13 cells transfected with Gm6135 siRNA and miR-203 inhibitor was measured by qRT-PCR (f) and western blot (g). The data are presented as mean ± standard deviation. *p < 0.05, according to Student’s t test. NC, negative control; qRT-PCR: quantitative reverse transcription-polymerase chain reaction; TLR4: TLR4: toll-like receptor 4 [Color figure can be viewed at wileyonlinelibrary.com]
3.6 MiR-203 suppresses esophageal squamous carcinoma cell (ESCC) cell proliferation
Our study shows that miR-203 mimics can effectively reduce the proliferation of SV40-MES-13 cells. However, overexpression of TLR4 overcame the inhibition of miR-203 mimics (Figure 6a). In contrast, miR-203 inhibitors promoted cell proliferation and were also abolished by TLR4 siRNA (Figure 6b).
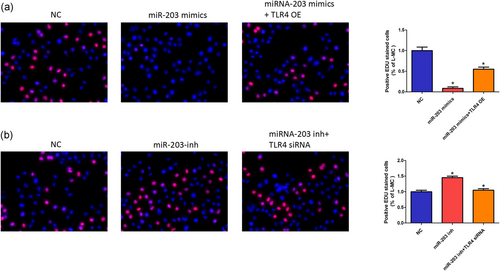
MiR-203 suppresses ESCC cell proliferation. (a,b) SV40-MES-13 cells were transfected with NC, miR-203 mimics, TLR4 OE, miR-203 inhibitor, and TLR4 siRNA as indicated. EDU images as shown in left panel (original magnification, ×200); quantitative number of positive EDU stained cells are shown in the right panel. The data are presented as mean ± standard deviation. *p < 0.05, Student’s t test. EDU: 5-Ethynyl-2’-deoxyuridine; ESCC: esophageal squamous carcinoma cell; NC: negative control; siRNA: small interfering RNA; TLR4: toll-like receptor 4 [Color figure can be viewed at wileyonlinelibrary.com]
4 DISCUSSION
Although many studies have confirmed the relationship between lncRNA and DN (C.P. Li, Wang, Wang, Song, & Liu, 2017; Reddy et al., 2014), the specific role of a single lncRNA in DN still needs further study. We used the early biochip data to find Gm6135 with abnormal expression associated with DN. In this experiment, the relative expression level of Gm6135 in kidney and high-glucose-cultured SV40-MES-13 cells of DN mice was confirmed by qRT-PCR, which also proved the accuracy of the chip data, further indicating that hyperglycemia plays an important role in the functional and structural abnormalities of DN. Subsequently, we demonstrated through EDU and CCK8 experiments that overexpression of Gm6135 resulted in abnormal proliferation of SV40-MES-13 cells. We all know that excessive proliferation of mesangial cells is closely related to the occurrence of DN. Therefore, this result suggests that there is a certain relationship between the abnormal expression of Gm6135 in the kidney and the development of DN.
Studies have shown that inflammatory response plays an important role in the early development of DN (Vaishnudevi & Viswanathan, 2017). Therefore, finding a direct relationship between Gm6135 and inflammation is beneficial for us to better study the relationship between lncRNA and DN. After combining the data of the database to predict all possible target genes of Gm6135, and classifying the target genes related to inflammation through gene ontology (GO) annotation information, we found the most direct target genes for inflammation. Under such research, we found that there is a repeat sequence between Gm6135 and TLR4 and that the repeat sequence is perfectly paired with the miR-203 sequence. On the basis of this prediction, we further confirmed the predicted results by the reporter gene, RNA pull down, and other experiments, confirming that Gm6135/TLR4 gene can bind to miR-203, and Gm6135 can affect TLR4 expression by binding to miR-203. This result suggests that Gm6135 may be involved in the inflammatory response of DN through TLR4.
TLR4 belongs to the family of model receptors, which are highly conserved receptor families that recognize multiple types of pathogen-associated molecular patterns or damage-related molecular patterns, thus representing the first line of defense. In addition, TLR-mediated signaling can lead to Innate immune cell activation, which produces two effects, one of which is called expression and secretion cytokines (proinflammatory cytokine), such as TNF-α, IL-12, IL-6, and so on. These cytokines can induce inflammation and promote antigen presentation and promote the pattern change of Th1 or Th2 in T helper cells (Th). Second, TLR can induce the expression of a costimulatory molecule and initiate the production of a specific immune response. Recently, TLR4 has been widely demonstrated to be involved in the pathogenesis of diabetes (Yu, Bo, Villani, Spencer, & Fu, 2016; Zhu, Han, Yuan, Xue, & Pang, 2018). We confirmed by qRT-PCR and western blot that the relative expression level of TLR4 was significantly upregulated in diabetic mouse kidney and SV40-MES-13 cells. At the same time, overexpression of TLR4 can significantly reverse the decrease in cell proliferation induced by transfection of miR-203 mimics, indicating that TLR4 plays an important role in the development of DN.
In conclusion, we found Gm6135 negatively regulated relative expression of miR-203, stimulated expression of TLR4 and promoted cell proliferation of SV40-MES-13 cells. Our study proved that the Gm6135/miR-203/TLR4 axis played an important role in DN and provided a novel perspective for the pathogenesis of DN.
ACKNOWLEDGEMENT
This work was supported by the grants from the Foundation of Huai’an science and technology bureau project (HAS201610 to Feng Bai)
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.