Prognostic value of aberrantly expressed methylation gene profiles in lung squamous cell carcinoma: A study based on The Cancer Genome Atlas
Abstract
Currently, research on genome-scale epigenetic modifications for studying the pathogenesis of lung cancer is lacking. Aberrant DNA methylation, as the most common and important modification in epigenetics, is an important means of regulating genomic function and can be used as a biomarker for the diagnosis and prognosis of lung squamous cell carcinoma (LUSC). In this paper, methylation information and gene expression data from patients with LUSC were extracted from the TCGA database. Univariate and multivariate COX analyses were used to screen abnormally methylated genes related to the prognosis of LUSC. The relationship between key DNA methylation sites and the transcriptional expression of LUSC-related genes was explored. A prognostic risk model constructed by four abnormally methylated genes (VAX1, CH25H, AdCyAP1, and Irx1) was used to predict the prognosis of LUSC patients. Also, the methylation levels of the key gene IRX1 are significantly correlated with the prognosis and correlated with the methylation of the site cg09232937 and cg10530883. This study is based on high-throughput data mining and provides an effective bioinformatics basis for further understanding the pathogenesis and prognosis of LUSC, which has important theoretical significance for follow-up studies on LUSC.
1 INTRODUCTION
Lung cancer ranks first among all cancers in terms of incidence, and it is also the most important cause of cancer deaths all over the world (Torre et al., 2015). According to histological differences, lung cancer can be divided into non-small-cell lung cancer (NSCLC) and small-cell lung cancer (SCLC), of which NSCLC accounts for approximately 85%, and 30% of NSCLC cases can be classified as lung squamous cell carcinoma (LUSC; Piperdi, Merla, & Perez-Soler, 2014). The incidence of LUSC is high, and the prognosis is poor (5-year survival rate < 15%); LUSC causes approximately 400,000 deaths per year worldwide. Recent studies have shown that LUSC is closely related to smoking and is more common in men than in women (Kenfield, Wei, Stampfer, Rosner, & Colditz, 2008). Although the occurrence of LUSC can be prevented to a certain extent by regulating smoking (Meza, Meernik, Jeon, & Cote, 2015), the mechanism of occurrence and development of LUSC is still not fully characterized. In recent years, there has been great progress in molecular targeted therapies for LUSC (Drilon, Rekhtman, Ladanyi, & Paik, 2012; Lee et al., 2013; Sun et al., 2016). However, due to targeted drug resistance and the toxicity of treatment, some patients do not benefit from it. Therefore, to decrease mortality and improve the management of LUSC, detection and risk stratification of LUSC is urgently needed to identify new early diagnostic biomarkers and therapeutic targets. In the current study, we aimed to find effective potential molecular biomarkers for predicting survival in LUSC.
NSCLC poses a great threat to human health. Epigenetic abnormalities are one of the most important mechanisms in the development of lung cancer (Yang & Schwartz, 2011). The abnormal methylation of DNA is the most common and important modification in epigenetics and is an important means of regulating the function of the genome. Selective methylation and demethylation of genes to form specific tissue types and regulate the expression of genes during development and differentiation are considered to be major factors leading to the development of multiple tumors (Kulis & Esteller, 2010). The study of the relationship between methylation and lung cancer has become an area that has drawn more attention. Studies have suggested that the altered DNA methylation patterns in tumor tissues may silence tumor-suppressor genes and activate oncogenes through hyper/hypomethylation. For example, the hypermethylation of the promoter of the suppressor gene SOX17 was found to silence the expression of this gene in 60.2% of primary lung cancer samples, thereby eliminating the inhibition of cell proliferation in lung cancer (Yin et al., 2012). Identification of gene methylation abnormalities can explain the instability and redundancy of the lung cancer genome and provide the basis for targeted therapy or risk prediction.
The Cancer Genome Atlas (TCGA; Tomczak, Czerwińska, & Wiznerowicz, 2015) has created a genomic data analysis process that can effectively collect, select, and analyze human tissue for genome changes. The TCGA database provides analysis of high-throughput data for various genomic changes, including DNA methylation. Bioinformatics analysis along with patient diagnostics, treatment information, and tumor pathology information can be combined to advance oncology research and lay the foundation for improved cancer prevention, early detection, and treatment. In this study, LUSC-related abnormally methylated and expressed genes were identified based on TCGA data, and the expression of abnormally methylated genes and related differential genes in LUSC patients was clearly defined. We conducted a series of bioinformatics and survival analyses, screened for biomarkers associated with methylation, and constructed a prognostic risk model to help us to predict the prognosis of LUSC patients. These results could provide new insight into the molecular mechanisms based on methylation in LUSC.
2 MATERIALS AND METHODS
2.1 Data processing and differential expression analysis
The TCGA Data Portal was terminated, and all TCGA data were transferred to the newly established Genomic Data Commons (GDC, https://gdc.cancer.gov/). Therefore, the original TCGA sequencing data related to abnormally methylated genes in LUSC patients in this study was obtained from the GDC, and the methylation data were obtained using Human Methylation 450k Illumina Infinium methylation arrays; the data included 42 normal samples, 370 LUSC samples, and related differentially expressed gene data, including 49 normal samples and 502 LUSC samples; due to the difference in the sample sizes of the two groups of data, the original data that had been preserved for the same sample and had corresponding survival information and status were analyzed. R software and related R packages were used to normalize and analyze the downloaded data to obtain abnormally methylated genes and differentially expressed genes. The relevant data TCGA provided is publicly available and open-ended; therefore, approval by a local ethics committee was not needed.
2.2 Protein–protein interaction (PPI) network construction and abnormally methylated genes function enrichment
As a search tool for searching protein–protein function relevance, the STRING (Szklarczyk et al., 2015; http://string-db.org) database, utilizes literature content to extract PPIs, we used STRING to characterize the PPI network of the genes, and a confidence score >0.4 was set as the cut-off criterion. Then, we used Cytoscape software (http://cytoscape.org/development_team.html) to visualize the resulting PPI network (Shannon et al., 2003).
The Cytoscape plug-in (BinGo) provides a comprehensive set of functional annotation tools for investigators to understand the biological meaning behind Gene Ontology (GO) and Kyoto Enclarge lists of genes. To further understand the function of these abnormally methylated genes, functional enrichment analyses were performed using the Cytoscape plug-in BinGo. The GO terms were selected with p < 0.05 as the cut-off condition.
2.3 Screening for prognosis-relevant signatures and risk score calculation
2.4 Joint survival analysis and correlation analysis
Combined survival analysis was performed by combining the screened, independently prognostic, abnormally methylated genes with the corresponding gene expression data. The joint survival curve was drawn through the survival R package. The resulting prognostically relevant genes were considered key genes for LUSC because their methylation levels and gene expression levels were significantly correlated with prognosis. Since aberrant methylation was thought to be associated with gene expression, to further explore the relevance, we extracted relevant sites for methylation of key genes from the downloaded LUSC methylation data. Next, we evaluated the correlation between key gene methylation sites and gene expression.
3 RESULTS
3.1 TCGA data analysis and construction of the PPI network
A total of 370 samples of methylated LUSC and 42 control samples were included in our analysis. Based on the relevant LIMMA software package (http://bioconductor.riken.jp/packages/3.0/bioc/html/limma.html), the abnormal methylation expression data from TCGA were extracted and analyzed. To narrow down the data range, we used |log FC| > 1, p < 0.05 as the screening conditions to obtain 211 abnormally methylated genes (Figure 1). To further investigate the potential links between these abnormally methylated genes, the STRING online database was used to mine and describe the PPI network, and a confidence score >0.4 was used as the cut-off criterion. Then, we used the Cytoscape software to visualize the generated PPI network; a total of 103 abnormally methylated genes were filtered into the complex PPI network, which contained 103 nodes and 225 edges (Figure 2), 118 of the 211 genes did not fall into the PPI network. Those genes were used for further functional and survival analysis to explore functional abnormalities caused by the aberrant methylation of genes in normal and diseased patients.
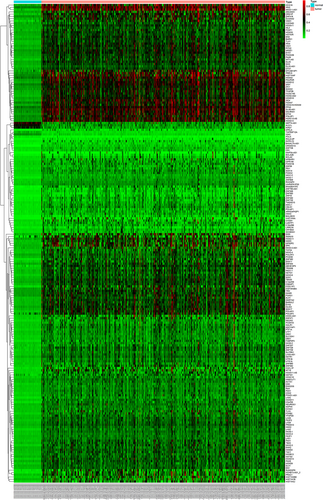
Heat maps of LUSC-related aberrant methylated genes. The color from green to red shows a trend from low expression to high expression. LUSC: lung squamous cell carcinoma [Color figure can be viewed at wileyonlinelibrary.com]
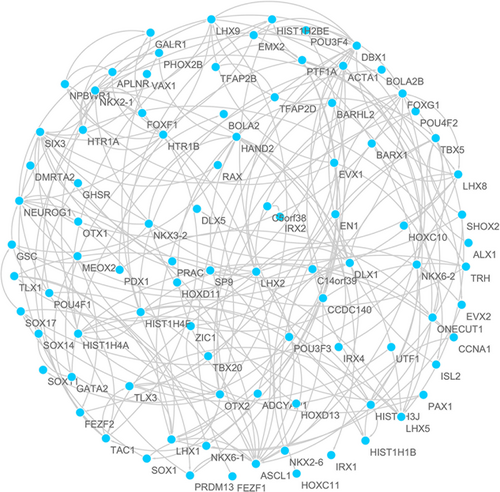
The PPI network of aberrant methylated genes in LUSC. LUSC: lung squamous cell carcinoma; PPI: protein–protein interaction [Color figure can be viewed at wileyonlinelibrary.com]
3.2 Functional enrichment analysis of abnormally methylated genes
To further understand the functional role of abnormally methylated genes in LUSC, we used the BinGo plug-in for functional enrichment analysis of these genes. The results showed that the abnormally methylated genes were particularly rich in molecular function (MF), biological processes (BP), and cell components classification. In the BP group, these genes were mainly enriched in regulation of DNA-dependent transcription, the RNA metabolic process, gene expression, etc. (Figure 3a). The MF is mainly enriched in multiple binding, such as chromatin DNA binding, DNA regulatory region binding, transcription regulator activity, and transcription factor binding (Figure 3b). Also, cellular component (CC) terms are mainly involved in the cytoplasm and nucleoplasm (Figure 3c).
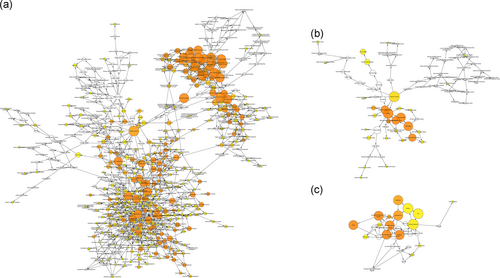
Gene ontology analysis of aberrant methylated genes associated with LUSC. Enrichment analysis of (a) biological processes; (b) molecular function; and (c) cellular component. LUSC: lung squamous cell carcinoma [Color figure can be viewed at wileyonlinelibrary.com]
3.3 Prognostic assessment of abnormally methylated genes and clinical features
We conducted a univariate Cox regression between abnormally methylated genes in LUSC patients, and the results showed that a total of 39 genes with p < 0.05 were greatly associated with OS (Table 1). And then, multivariate Cox regression was applied to construct a prognostic risk model. We found that four genes (ventral anterior homeobox 1 [VAX1], cholesterol 25-hydroxylase [CH25H], adenylate cyclase activating polypeptide 1 [AdCyAP1], and iroquois homeobox 1 [Irx1]) were independent prognostic indicators for LUSC (Figure 4), and the risk model constructed by them could be used for prognosis assessment. The risk score was input as follows: (−1.973 × expression level of ADCYAP1) + (−1.124 × expression level of IRX1) + (1.384 × expression level of VAX1) + (−1.589 × expression level of CH25H). Also, based on the cut-off of the median risk score, a total of 405 patients were classified into a high-risk group (n = 202) and a low-risk group (n = 203) according to the median risk score. The results of survival analysis showed that patients in the high-risk group have a significantly worse OS than that of patients in the low-risk group (p = 3e−05, Figure 5a). Then, we used time-dependent ROC curves to evaluate the prognostic ability of risk model. The area under the curve (AUC) for the four-biomarkers prognostic model was 0.644 at 5 years of OS (Figure 5b).
| Gene | HR | Z | p |
|---|---|---|---|
| GHSR | 0.226142 | −2.83672 | 0.004558 |
| ADCYAP1 | 0.206839 | −2.75709 | 0.005832 |
| TRIM58 | 0.271923 | −2.70983 | 0.006732 |
| ZNF454 | 0.237009 | −2.70699 | 0.00679 |
| CH25H | 0.262343 | −2.66953 | 0.007596 |
| COX11P1 | 0.3957 | −2.64223 | 0.008236 |
| ZNF730 | 0.206126 | −2.57848 | 0.009924 |
| PCDH8 | 0.230702 | −2.55218 | 0.010705 |
| IRX1 | 0.212528 | −2.50359 | 0.012294 |
| ZNF568 | 0.182196 | −2.45557 | 0.014066 |
| NKX2–3 | 0.311258 | −2.43929 | 0.014716 |
| EVX2 | 0.342645 | −2.37066 | 0.017756 |
| NKX2–6 | 0.338588 | −2.3206 | 0.020308 |
| SOX14 | 0.207648 | −2.32048 | 0.020315 |
| ZNF729 | 0.282189 | −2.31958 | 0.020364 |
| EMX2 | 0.250729 | −2.29638 | 0.021654 |
| CBLN1 | 0.268631 | −2.25892 | 0.023888 |
| ZNF790-AS1 | 0.27809 | −2.22204 | 0.026281 |
| C5orf38 | 0.289961 | −2.19936 | 0.027852 |
| FOXB2 | 0.336379 | −2.17058 | 0.029963 |
| ZNF69 | 0.219363 | −2.13281 | 0.03294 |
| SOX11 | 0.41728 | −2.12322 | 0.033735 |
| POU3F3 | 0.324792 | −2.11451 | 0.034472 |
| DBX1 | 0.240704 | −2.10174 | 0.035576 |
| ZNF492 | 0.273512 | −2.09055 | 0.036568 |
| ALX1 | 0.40646 | −2.07893 | 0.037624 |
| SOX1 | 0.33162 | −2.06098 | 0.039305 |
| ZNF728 | 0.320848 | −2.0513 | 0.040237 |
| ZNF257 | 0.308881 | −2.05121 | 0.040246 |
| ABCB10P4 | 0.413226 | −2.0358 | 0.04177 |
| ACTA1 | 0.244979 | −2.02704 | 0.042658 |
| NKX6–2 | 0.2789 | −2.016 | 0.0438 |
| PDX1 | 0.268326 | −2.00947 | 0.044488 |
| VAX1 | 0.373035 | −2.00838 | 0.044603 |
| GRAMD1A | 0.168909 | −2.00591 | 0.044866 |
| RAX | 0.227745 | −1.99582 | 0.045954 |
| TBX20 | 0.371405 | −1.99235 | 0.046332 |
| SOX17 | 0.432915 | −1.97682 | 0.048062 |
| ZNF808 | 0.187345 | −1.96339 | 0.049601 |
- Note. HR: hazard ratio; OS: overall survival.
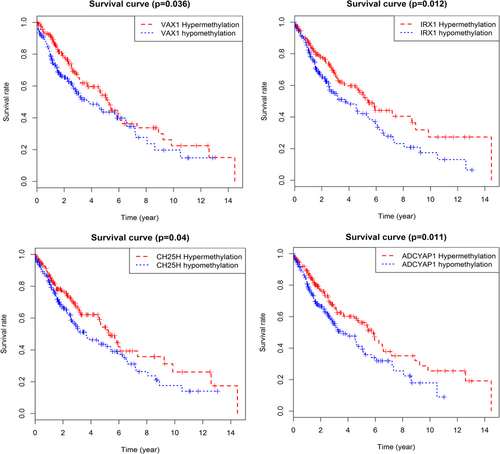
Kaplan–Meier survival curves of four independent prognostic factors [Color figure can be viewed at wileyonlinelibrary.com]
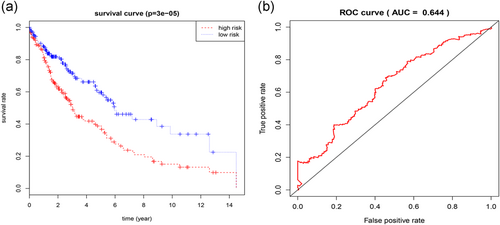
(a) Kaplan–Meier survival curves for overall survival outcomes according to the risk cut-off point. (b) Time-dependent ROC curves analysis for 5-year survival prediction by the four genes. ROC: receiver operating characteristic [Color figure can be viewed at wileyonlinelibrary.com]
3.4 Correlation analysis of differential methylation sites and gene expression
Using p < 0.05 as a meaningful screening criterion for combined survival, the methylation and gene expression levels of the prognosis-related gene IRX1 were significantly correlated with prognosis (Figure 6). Also, the prognosis genes methylation sites based on relevant data mining in TCGA are searched, and correlations between site and gene expression were analyzed, using |Cor| > 0.5 as a screening condition (Table 2). The gene expression of IRX1 and VAX1 was found to be related to the methylation level of multiple sites, and the gene IRX1 showed a negative correlation, while VAX1 showed a positive correlation (Figure 7).
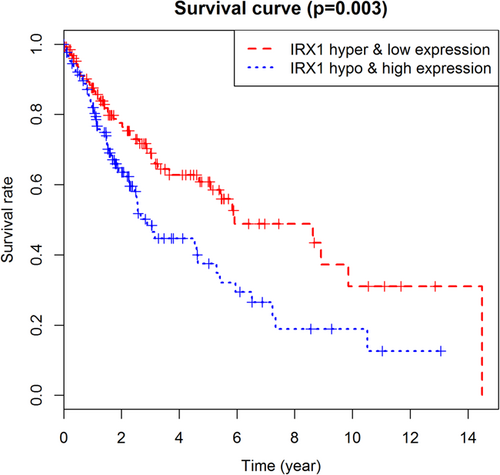
Kaplan–Meier survival curves for the combination of gene IRX1 methylation and expression. IRX1: iroquois homeobox 1 [Color figure can be viewed at wileyonlinelibrary.com]
| Methylation site | Correlation | p | |
|---|---|---|---|
| IRX1 | cg10530883 | −0.502 | 1.785e−25 |
| cg09232937 | −0.502 | 1.839e−25 | |
| VAX1 | cg10143067 | 0.556 | 6.501e−32 |
| cg18459489 | 0.57 | 7.532e−34 | |
| cg15668468 | 0.54 | 5.872e−30 | |
| cg03851159 | 0.569 | 8.79e−34 | |
| cg18709545 | 0.617 | 6.186e−41 |
- Note. IRX1: iroquois homeobox 1; VAX1: ventral anterior homeobox 1.
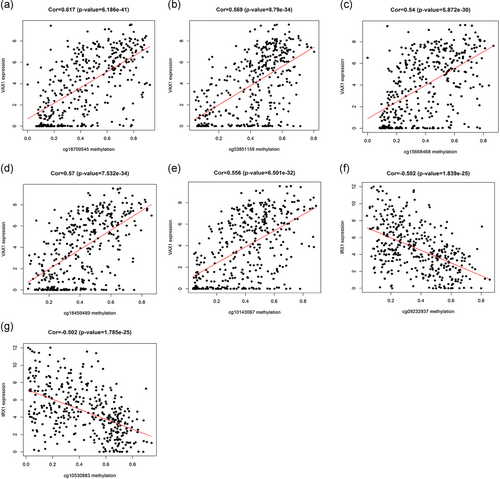
The relationship between gene expression and site methylation. (a–e) The gene expression of VAX1 and site methylation; (f,g) The gene expression of IRX1 and site methylation. IRX1: iroquois homeobox 1; VAX1: ventral anterior homeobox 1 [Color figure can be viewed at wileyonlinelibrary.com]
4 DISCUSSION
Lung cancer, which is mainly associated with smoking, is one of the most common malignancies in humans. Other factors, such as the relationship between viral infection and lung cancer, are currently hot topics. NSCLC is a huge threat to human health, and LUSC accounts for 30% of NSCLC. The study of methylation and its relation to LUSC is a field of growing interest. In recent years, with the clinical application of epidermal growth factor receptor tyrosine kinase inhibitor (EGFR-TKI), the therapeutic effect of lung adenocarcinoma has been significantly improved. At the same time, LUSC is being studied as an independent type of lung cancer to obtain new therapeutic targets. These studies have focused on changes in the molecular biology of LUSC and have made many advances. The rapid development of gene analysis technology provides the possibility for in-depth exploration of the molecular characteristics of LUSC and provides valuable evidence for prognostic evaluation and molecular targeted therapy.
Epigenetics has led to the discovery that not only cytogenetic changes but also epigenetic abnormalities, including DNA methylation, are involved in the pathogenesis of LUSC. Accumulating evidence has shown that DNA methylation, a major molecular mechanism of epigenetic changes, is correlated with human malignant tumors, including lung cancer (Chakravarthi, Nepal, & Varambally, 2016; Huang et al., 2015). Genes with aberrant DNA methylation can be noninvasive biomarkers for the detection and diagnosis of cancer (Chen et al., 2014; Gloss & Samimi, 2014; Luttmer et al., 2016). Therefore, to explore the molecular mechanism of LUSC progression and the epigenetic changes that determine new biomarkers, early diagnosis, treatment, and prognosis of LUSC is crucial. The independence and stability of abnormal DNA methylation analysis make it an effective method for identifying prognostic biomarkers (Dinardo et al., 2017; Marcucci et al., 2014; Ni, Ye, & Huang, 2017). Recent research results showed that the methylation of the P16, RASSF1A, APC, or SHOX2 genes were significantly associated with lung cancer and could serve as potential noninvasive biomarkers in lung cancer (Ni et al., 2017). Other studies have shown that TRIM58/cg26157385 methylation is associated with eight prognostic genes, including A2ML1 and CCNE1, in LUSC (W. Zhang et al., 2018). Also, some research also identified and validated the methylation of the HOXA9 promoter as a prognostic biomarker associated with high risk in Stage I lung adenocarcinoma patients (Robles et al., 2015). Thus, bioinformatics analysis of the functional enrichment and prognostic evaluation of abnormally methylated genes can provide clinicians with new tools that can be used to treat patients and predict prognoses.
In this study, we aimed to investigate abnormally methylated genes between LUSC patients and normal samples and to identify prognostic biomarkers related to methylation-mediated expression. We initially identified 211 abnormally methylated genes and 1,854 differentially expressed genes using the LIMMA software package. To gain insights into the functional roles of these LUSC aberrantly methylated genes, functional enrichment of these genes was performed. These genes were found to be involved in intracellular and nucleoplasm pathways and were mainly enriched related to the regulation of transcription, DNA-dependent processes, RNA metabolic processes and a variety of other functions, such as DNA regulatory region binding and transcription factor binding. These functional enrichment items not only clearly show the changes in gene function that may result from abnormal DNA methylation in different samples but also show the interaction of genes at the functional level.
In addition, through univariate and multivariate Cox analyses of the abnormally methylated genes, it was found that the prognostic risk model (high risk and low risk) which was constructed by the four abnormally methylated genes (VAX1, CH25H, ADCYAP1, and IRX1) could predict the survival rate, and each of these four genes was an independent prognostic factor for LUSC. The predictive power of the risk model was evaluated by a time-dependent ROC curve, and the result of AUC was more than 0.6. Despite its limitations, it could be concluded that the prognostic model we constructed had a certain accuracy and sensitivity in assessing the prognosis of patients. As a transcription factor, methylated VAX1 is thought to be highly correlated with bladder cancer (BC) recurrence and may serve as a useful biomarker for predicting BC recurrence (Zhao et al., 2012). Also, VAX1 were methylated in more than 80% of the adenocarcinomas, which is including lung adenocarcinoma (Rauch et al., 2012). However, there are few studies on the methylation of VAX1 in LUSC. In this study, hypermethylation of VAX1 showed a high survival rate and could be used as one of the prognostic indicators of LUSC. And the expression of VAX1 gene was positively correlated with the methylation levels of multiple sites. This may be because the hypermethylation of VAX1 enhances the expression of the gene and improves the patient’s quality of life. Of course, this requires further research to confirm our result. CH25H is a gene that codes for a cholesterol 25-hydroxylase involved in cholesterol and lipid metabolism (Bauman et al., 2009). Studies have shown that downregulation of CH25H can reduce plasma cell responses after immune challenge (Hannedouche et al., 2011). Also, low expression of CH25H is closely associated with a poor prognosis in breast cancer (Mittempergher et al., 2013) and may be a potential target for distant metastasis control in breast cancer. We have found that the downregulation of CH25H expression in LUSC patients may shorten the survival time by reducing the patient’s immune response, which requires further verification through experiments. CH25H, which has been less reported in lung cancer, may serve as a new marker for further study of LUSC. The ADCYAP1 gene is involved in various biological processes, including cell growth, proliferation, and differentiation (Wolman, Heppner, & Wolman, 1997). Research showed that ADCYAP1 hypermethylation is closely related to cervical cancer and could be used as an effective and sensitive methylation biomarker for the early diagnosis of cervical cancer (Jung et al., 2011). Also, gene expression of ADCYAP1 is associated with multiple cancers, such as neuroblastoma (Isobe et al., 2004) and breast cancer (García-Fernández et al., 2004). Pituitary adenylate cyclase-activating polypeptide, a protein encoded by gene ADCYAP1, can be used as a potential serum marker for non-small-cell lung cancer (H. Zhang, Chen, & Huang, 2012). In this study, compared with hypomethylation, hypermethylation showed a high survival rate, and methylation abnormalities affected the expression of the ADCYAP1 gene and affected LUSC-related biological processes, such as positive regulation of cell proliferation and cell–cell signalling.
IRX1, a member of the Iroquois homeobox family of transcription factors, was identified as a potential tumor-suppressor gene in head and neck squamous cell carcinoma (Bennett et al., 2008). In this study, IRX1 expression was downregulated and showed hypermethylation. Correlation analysis showed that the expression level was negatively correlated with the methylation level of the site cg09232937 and cg10530883. This may be due to the hypermethylation of the sites leading to IRX1 transcriptional silencing, which results in dysregulation of the expression, which in turn affects the progression of the disease and the prognosis of the patient. Overexpression of Irx genes may cause lung dysplasia by inducing lung dysmorphogenesis with thickened mesenchyme (Doi, Lukosiute, Ruttenstock, Dingemann, & Puri, 2011), in which case it can further lead to lung cancer. Moreover, the hypomethylation of IRX1 is associated with a high risk of lung metastasis (Lu et al., 2015). In our study, we found that the survival rate of patients with hypermethylation of the four genes was higher than that of patients with hypomethylation of those genes, and p < 0.05 was statistically significant. A combined analysis of gene expression and methylation levels revealed that the survival rate of patients with hypermethylation/low gene expression of IRX1 was significantly higher than that of patients with hypomethylation/high gene expression of IRX1. Hypermethylation often leads to transcriptional silencing and chromosome instability, causing gene dysregulation and cell differentiation disorder. Also, the expression of IRX1 was negatively correlated with the hypermethylation level of the site cg09232937 and cg10530883, which suggested that DNA hypermethylation may be an important reason for the downregulation of IRX1 expression in LUSC patients. These results provide a basis for bioinformatics and a related theoretical basis for further experimental validation.
5 CONCLUSION
Identification of differentially expressed genes is widely used in molecular biology studies to obtain markers of cancer diagnosis, treatment, and prognosis. In conclusion, in this study, we performed whole-genome methylation analysis of LUSC patients based on the TCGA database and found some abnormally methylated genes related to the development of LUSC. Univariate and multivariate Cox regression analysis showed that the prognostic risk model constructed by four abnormally methylated genes (VAX1, CH25H, ADCYAP1, and IRX1) proved to be an independent prognostic factor for LUSC. Also, the expression and methylation levels of the key gene IRX1 are significantly correlated with prognosis and are negatively correlated with the methylation of the site cg09232937 and cg10530883. Although experimental verification is required, our study can provide a basis for future diagnosis, treatment, and prognosis of LUSC. Also, our research has important theoretical significance in guiding follow-up studies of LUSC.
ACKNOWLEDGMENTS
This study is supported by the grants from National Natural Science Foundation of China (81673799) and National Natural Science Foundation of China (81703915).
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.