NONO ubiquitination is mediated by FBW7 and GSK3 β via a degron lost upon chromosomal rearrangement in cancer
Abstract
NONO is an RNA-binding protein involved in transcription, mRNA splicing, DNA repair, and checkpoint activation in response to UV radiation. NONO expression has been found altered in several tumor types, including prostate, colon, breast, melanoma, and in papillary renal carcinoma, in which an X chromosome inversion generates a NONO-TFE3 fusion protein. Upon such rearrangement, NONO loses its C-terminal domain. Through bioinformatics analysis, we identified a putative degron motif, known to be recognized by the Skp1-Cul1-F-box-protein (SCF) complex. Here, we evaluated how this domain could affect NONO protein biology. We showed that NONO interacts with the nuclear FBW7α isoform and its ubiquitination is regulated following modulation of the GSK3β kinase. Mutation of T428A/T432A within the degron impaired polyubiquitination upon FBW7α and GSK3β overexpression. Overall, our data suggest that NONO is likely subjected to proteasome-mediated degradation and add NONO to the list of proteins targeted by FBW7, which is itself often deregulated in cancer.
1 INTRODUCTION
NONO is an abundant and highly conserved nuclear protein involved in a wide range of cellular processes (Shav-Tal & Zipori, 2002). Human NONO, together with PSF/SFPQ and PSPC1, belongs to the DBHS (Drosophila behavior human splicing) protein family (Kowalska et al., 2012). Its gene, located on chromosome Xq13.1, encodes a 471 amino acid protein containing two RNA recognition motifs (RRMs) and a helix-turn-helix domain followed by a series of charged amino acids that likely form a DNA-binding unit (Dong et al., 1993). Together with PSF/SFPQ and PSPC1, NONO is a core protein component of paraspeckles, organelles that control gene expression through the nuclear retention of RNA species containing double-stranded regions that are subjected to A-to-I-editing and contribute to cell reprogramming upon differentiation, viral infection, or stress cues (Fox & Lamond, 2010). In addition to its nucleic acid binding properties, NONO has been shown to interact with the carboxyl-terminal domain of the largest subunit of RNA polymerase II, suggesting a role in coupling pre-mRNA synthesis and processing (Emili et al., 2002). NONO is a multifunctional protein playing a role in many cellular pathways. It is involved in the circadian clock control through the interaction with the PERIOD protein, regulating the p16-Ink4A expression (Kowalska et al., 2013). Moreover, NONO induces radio-resistance in response to ionizing radiations and it is described to regulate the non-homologous end joining (NHEJ) process (Krietsch et al., 2012; Li et al., 2009). NONO was described to be involved in the regulation of the DNA damage response to UV radiation (Alfano et al., 2016a). NONO expression is altered in several tumor types, including prostate (Ishiguro et al., 2003), colon (Nelson et al., 2012), breast (Pavao et al., 2001), and melanoma (Schiffner, Zimara, Schmid, & Bosserhoff, 2011). Interestingly, NONO has also been found altered at the chromosomal level in papillary renal carcinoma. In particular, it is rearranged with the TFE3 gene, encoding a transcription factor, following an Xp11.2 translocation, which generates the fusion protein NONO-TFE3. Fewer than 10 cases with the NONO-TFE3 rearrangement are described in the literature, nonetheless it seems that this chromosomal aberration is a driver of carcinogenesis. Indeed the fusion protein has been hypothesized to act as an activator of the promoter of MET encoding a receptor tyrosine kinase, which is aberrantly activated in multiple cancer types (Clark et al., 1997). Also other rearrangements, involving the TFE3 gene, were described suggesting the relevance of this molecular rearrangement in the development of papillary renal carcinoma (Kauffman et al., 2014). Upon the chromosomal rearrangement, NONO loses a small DNA portion in its C-terminal domain (Clark et al., 1997). Here, we set out to evaluate a possible role of this domain in NONO protein biology, which might have an impact in the context of tumor development.
2 MATERIALS AND METHODS
2.1 Cell culture, transfection, and reagents
The embryonic kidney HEK293 cell line was purchased from the American Type Culture Collection (ATCC, Manassas, VA). Cells were cultured in DMEM (Dulbecco's Modified Eagle Medium) (Thermo Fisher Scientific, Carlsbad, CA) supplemented with 10% foetal bovine serum (Thermo Fisher Scientific), penicillin (100 U/ml), streptomycin (100 µg/ml), and 2 mM glutamine at 37°C in 5% CO2. The MG132 proteasome inhibitor (30 µM) was purchased from Santa Cruz Biotechnology, Dallas, TX, dissolved in dimethyl sulfoxide (DMSO) and stored at −20°C in small aliquots. MG132 efficacy in blocking the proteasome was evaluated assessing through Western blot the increase of total protein ubiquitination. Transfections were carried out by using Xtreme-GENE 9 (Sigma–Aldrich, St Louis, MO) according to the manufacturer instructions. HA GSK3 beta wt pcDNA3 was a gift from Jim Woodgett (Addgene plasmid # 14753). The other plasmids used were obtained upon cloning of each respective cDNA obtained through RT-PCR with the primers indicated in Supplementary Table S1. Site directed mutagenesis of the degron motif was carried out through the QuikChange Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA) according to the manufacturer instructions with the primers listed in Supplementary Table S1.
2.2 Western blotting
Cells were lysed with the JS lysis buffer (50 mM N-2-idrossietilpiperazin acid-N′-2-ethanesulfonic acid [HEPES] pH 7.5, 1% [v/v] TritonX-100, 150 mM NaCl, 5 mM EGTA) containing phosphatase inhibitors (50 mM NaF, 20 mM sodium pyrophosphate, 1 mM sodium vanadate) and protease inhibitors (2 mM phenylmethylsulfonyl fluoride [PMSF] and 1 mg/ml aprotinin). Lysates containing equal amounts of proteins, estimated through the Bradford assay (Bio-Rad, Munchen, Germany), were subjected to SDS-page and Western blot with the following antibodies: polyclonal anti-p54 nrb-(C-17) detecting NONO, anti-GAPDH, anti-HA tag, and the anti-mouse (1:3000) and anti-rabbit (1:3000) secondary antibodies coupled to horseradish peroxidase, which were purchased from Santa Cruz Biotechnology; the monoclonal anti-HA tag (clone 12CA5), which was purchased from Roche, Mannheim, Germany; the anti-p54 monoclonal antibody (clone 78.1) detecting NONO, which was purchased from Millipore, Burlington, MA.
2.3 Immunoprecipitation assay
Cells were lysed with the JS lysis buffer containing phosphatase and protease inhibitors, as described above. Each immunoprecipitate was performed with 2 mg of total protein lysate. Pre-clearing was performed with protein G (Thermo Fisher Scientific) previously washed and equilibrated with the lysis buffer, for 45 min rocking at 4°C. Thereafter, the samples were centrifuged for 5 min at 1,000g and the supernatant was incubated with 2 μg of antibody rocking over night at 4°C. Then protein G was added for 45 min rocking at 4°C to isolate the immune complexes. Subsequently, the lysate was centrifuged for 5 min at 1,000g and the pellet washed five times with the JS lysis buffer. The samples were resuspended in 2× SDS Laemmly buffer and boiled at 99°C for 10 min followed by SDS polyacrylamide gel electrophoresis.
2.4 Pulse-chase assay
HEK293 cells were incubated with depletion medium—Met-Cys free DMEM (Dulbecco's Modified Eagle Medium, no methionine, no cysteine) (Thermo Fisher Scientific)—for 30 min. For the pulse phase, 0,35 ml of [35S]-Meth (PerkinElmer, Billerica, MA) working solution was added to cells and left for 40 min. Before adding the “chase” medium, (complete DMEM medium supplemented with 10 mM HEPES, 5 mM cysteine, 5 mM methionine); cells were washed to completely remove the radioactive compound. Cells were collected after 28, 30, and 32 h of treatment. [35S]-Meth incorporation was determined by TCA precipitation (Link & LaBaer, 2011).
3 RESULTS
3.1 Identification of a putative degron motif within NONO and analysis of NONO half-life
The chromosomal inversion described in papillary renal cell carcinoma, affecting NONO and TFE3, causes in both the loss of a protein sequence (Clark et al.). To investigate the function of NONO C-terminal domain and gain insight into how its loss could affect NONO biology, we used the “ELM” (http://elm.eu.org/) program for the prediction of linear motifs in order to search for the presence of conserved domains. Upon bioinformatical analysis, we identified a putative “degron” motif common to many oncoproteins which are targeted for degradation by the Skp1-cullin-F-box-protein (SCF) complex (Figure 1a). So, we set out to evaluate whether NONO turnover could be degron-dependent.
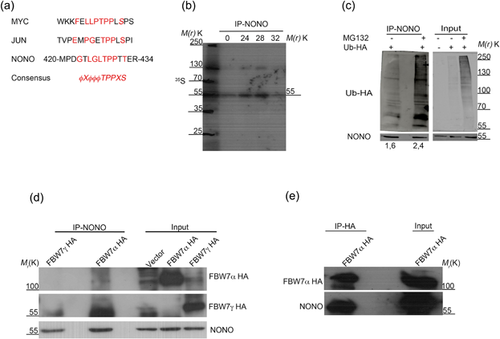
First, we tried to determine NONO half-life in HEK293 cells. Owing to the long half-life we could not detect protein degradation through cycloheximide because the long treatment with the drug was toxic to the cells (data not shown). So, we incubated cells in a depletion medium (devoid of methionine and cysteine) for 30 min at 37°C, followed by labeling through [35S]-methionine precursors for 40 min. At the end of labeling, HEK293 cells were washed and incubated with an excess of cold precursor. Cells were then harvested at the indicated time points and NONO was immunoprecipitated from the total protein lysates. Autoradiography analysis showed that NONO protein half-life was approximately 32 h (Figure 1b), which is consistent with in silico predictions achieved through different software (such as http://web.expasy.org/protparam/ and http://protein-n-end-rule.leadhoster.com/).
3.2 NONO is polyubiquitinated upon FBW7α interaction
To assess whether NONO can undergo polyubiquitination followed by degradation through the proteasome (Welcker, Orian, Grim, Eisenman, & Clurman, 2008), HEK293 cells were transfected with a plasmid expressing an HA-tagged ubiquitin and after 36 h treated with the proteasome inhibitor MG132 for 12 additional hours. Western blot analysis following immunoprecipitation showed that NONO was polyubiquitinated and the inhibition of proteasome activity increased the amount of polyubiquitination (Figure 1c). To investigate a possible role of the SCF complex in NONO ubiquitination, we analyzed whether NONO could interact with the F-box and WD repeat domain-containing 7 protein (FBW7), an ubiquitin ligase part of the SCF complex involved in the degradation of many proteins, which interacts with its target substrates through a Cdc4 phosphodegron (CPD) motif favoring polyubiquitination and degradation (Welcker et al., 2008). So, we transfected HEK293 cells with plasmids expressing the two nuclear FBW7 isoforms (either FBW7α or FBW7γ) fused to the HA-tag (Welcker, Orian, Grim et al., 2004). Upon 36 h of transfection, cells were treated with MG132 and harvested after 12 h. Western blot analysis following immunoprecipitation with anti NONO antibody, showed the specifical interaction with the FBW7α isoform (Figure 1d). To further confirm the interaction between the two proteins, we transfected and treated HEK293 cells in the same previous condition, followed by immunoprecipitation of HA-antibody (Figure 1e). Indeed, NONO was found to interact reciprocally with FBW7α.
To evaluate a possible role of FBW7α in the ubiquitination of NONO, we co-transfected HEK293 cells with plasmids expressing FLAG-tagged FBW7α and HA-tagged ubiquitin, and upon 36 h of transfection, we treated cells with MG132 for 12 additional hours. Western blot analysis following immunoprecipitation of the endogenous NONO showed that the overexpression of the FBW7α isoform increases NONO polyubiquitination (Figure 2a). Collectively, these data demonstrate that NONO ubiquitination relies on its interaction with the FBW7α protein.
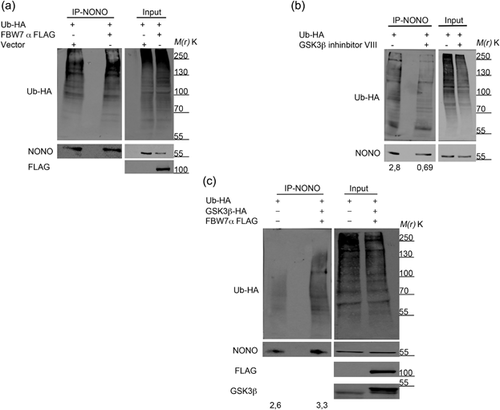
3.3 NONO polyubiquitination is triggered by GSK3β-mediated phosphorylation of the degron motif
The ability of FBW7 to interact with its target depends on a priming phosphorylation of the degron motif through the GSK3β kinase (Welcker, Orian, Jin et al., 2004). To evaluate a possible role of GSK3β in the regulation of NONO polyubiquitination, we transfected HEK293 cells with a plasmid expressing HA-tagged ubiquitin. Upon 36 h of transfection, we treated HEK293 cells with MG132 for 4 h followed by co-treatment with the GSK3β inhibitor VIII for 8 additional hours. We found that the pharmacological block of GSK3β activity effectively reduced NONO ubiquitination (Figure 2b). To further evaluate the effect of GSK3β modulation on NONO, we overexpressed in HEK293 both HA-tagged GSK3β and FLAG-tagged FBW7α. Western blot analysis following NONO immunoprecipitation showed that, as expected, the overexpression of GSK3β along with FBW7α induced an increase of NONO ubiquitination (Figure 2c).
In order to test whether the degron motif could be responsible for NONO ubiquitination, we mutated the two threonine residues (T428A/T432A) within NONO degron, which represent the predicted phosphorylation sites in the degradation consensus of many SCF target proteins. Upon immunoprecipitation of either FLAG-tagged NONO wt or its mutant form, which were co-transfected with both FBW7α and HA-tagged ubiquitin, we observed a reduced polyubiquitination of T428A/T432A protein with respect to the control (Figure 3a), demonstrating the involvement of the degron motif in NONO ubiquitination.
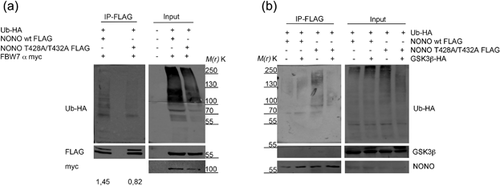
Finally, we analyzed the effect of GSK3β overexpression onto the NONO degron mutant. We co-transfected in HEK293 HA-tagged ubiquitin along with FLAG-tagged NONO wt or its mutant form and the GSK3β kinase. We found that, whereas GSK3β kinase overexpression was able to increase the ubiquitination of NONO wt, the mutated form showed a reduced ubiquitination, consistent with an inability of the kinase to phosphorylate the degron motif. Collectively, these data demonstrated the involvement of the SCF complex in NONO polyubiquitination through the recognition of the degron motif, which is phosphorylated by the GSK3β kinase.
4 DISCUSSION
NONO is a multifunctional protein involved in many biological processes, most of them occurring in the nucleus (Shav-Tal & Zipori, 2002). Recently, we showed that NONO is involved in the DNA damage response following UV radiation exposure suggesting that its mysfunction could contribute to tumorigenesis (Alfano et al., 2016a, 2016c). Consistently, NONO expression is altered in several tumor types, including prostate (Ishiguro et al., 2003), colon (Nelson et al., 2012), breast (Pavao, Huang, Hafer, Moreland & Traish, 2001), and melanoma (Schiffner et al., 2011). At present, over hundred missense, nonsense, or frameshift NONO point mutations have been found in cancer, according to the catalog of somatic mutations in cancer (COSMIC, http://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=NONO). The only molecular alteration affecting NONO at the chromosomal level reported so far, occurred in papillary renal cell carcinoma generating the NONO-TFE3 fused protein (Kauffman et al., 2014). During the molecular inversion NONO loses an amino acidic portion in its C-terminal domain (Clark et al., 1997). Here, we set out to evaluate the physiological role of such region within NONO protein biology and gain insight into how alterations of this region could potentially underlie tumorigenesis. Through a bioinformatical analysis we identified a putative degron motif, which is responsible for the degradation of many proteins by the SCF complex (Ang & Wade Harper, 2005). The SCF complex, along with the anaphase-promoting complex/cyclosome (APC), are among the main cellular machineries regulating protein turnover within the ubiquitin-proteasome pathway. They control cell cycle progression and mitotic exit acting on the stability of the cyclin-dependent kinases (Peters, 1998). The first step in this type of intracellular protein degradation is the protein target polyubiquitination by different enzymes, named E1, E2, and E3, which recognize and catalyze the conjugation of the ubiquitin molecules to the amino group of a lysine residue of the target protein, generating long ubiquitin chains. Polyubiquinated proteins are recognized and degraded into small peptides by the 26S proteasome complex (Reinstein & Ciechanover, 2006). Inappropriate ubiquitin-mediated protein degradation has been implicated in a number of pathological conditions including certain forms of cancer (Shen, Schmitt, Buac, & Dou, 2013; Takalo, Salminen, Soininen, Hiltunen, & Haapasalo, 2013). Target recognition depends on the action of various proteins, including FBW7, which has three isoforms (named α, β, and γ), deriving from alternative splicing (Spruck et al., 2002). FBW7α and FBW7γ proteins localize in the nucleus and nucleolus respectively, whereas FBW7β is present in the cytoplasm (Welcker, Orian, Grim et al., 2004). FBW7 controls protein fate of many cell cycle and growth regulators, including key cancer driver genes, such as cyclin E, MYC, JUN, and Notch (Welcker, Orian, Grim et al., 2004), and indeed acts as a tumor suppressor that is frequently inactivated in cancer (Welcker et al., 2008). In this context, we analyzed whether NONO turnover, physiologically, might be regulated through the SCF degradation pathway, which could contribute to clarify NONO role in various cancer types. To understand the possible function of the identified degron motif, which is lost upon the NONO-TFE3 rearrangement, we first assessed NONO half-life showing, through a pulse and chase labeling, that it corresponds to approximately 32 h (Figure 1b), which is consistent with in silico predictions and positions NONO in the class of long-lived proteins (Toyama & Hetzer, 2013). NONO localizes in the nuclear compartment (Shav-Tal & Zipori, 2002) and consistently, we demonstrated, through reciprocal co-immunoprecipitations, a specific interaction with the α isoform of FBW7 (Figure 1c), which is responsible for target recognition by the SCF complex (Welcker et al., 2008). Treatment with MG132 precluded co-localization analysis through immunofluorescence because cell detached from slides. The SCF complex acts as an ubiquitin conjugator favoring target degradation through the proteasome (Welcker et al., 2008) and in fact we detected accumulation of polyubiquitinated NONO, upon inhibition of the proteasome, which increased following overexpression of FBW7α (Figures 1c and 2a). Since FBW7-mediated target ubiquitination depends on the phosphorylation of serine or threonine residues within the degron motif by GSK3β (Xu, Kim, & Gumbiner, 2009), we assessed the effects of GSK3β modulation (either of its inhibition or overexpression) on NONO ubiquitination (Figures 2b and 2c). As expected NONO ubiquitination depended on the GSK3β modulation and it was inhibited by mutation of the phosphorylable threonine residues within the degron motif even upon ectopic overexpression of both FBW7α and GSK3β (Figures 3a and 3b), suggesting that NONO could be a direct target of GSK3β-phosphorylation although a shift of NONO weight through Western blot was not observed.
Collectively, these data identify NONO as a novel target of FBW7α and GSK3β shedding light on its possible degradation mechanism, which, as we showed, is likely dependent on the SCF complex activity. Although we demonstrated the involvement of NONO “degron” on the protein polyubiquitination, we never observed a change in NONO abundance upon MG132 treatment, probably owing to the long protein half-life.
These data are relevant not only for as concerns the physiological role of NONO in “normal” conditions (in particular it will be interesting to analyze these findings with respect to those reporting a role of NONO in the regulation of cell cycle and the circadian rhythms (Kowalska et al., 2013), but also in the cancer context. First, it is possible that the N-terminal region of NONO, within the NONO-TFE3 chimera, is aberrantly expressed in time owing to the lack of its degron motif, although further studies will be necessary to determine the oncogenic role of this fusion protein in renal cell carcinomas. Moreover the loss of NONO C-terminal domain in the NONO-TFE3 chimera, depletes also a region of NONO probably involved in the regulation of protein-protein interactions (given to the presence of a proline rich domain) whose loss could underlie tumorigenesis through other mechanisms.
Then, it is likely that these findings will help to clarify the roles of NONO in cell proliferation and cancer, which are still poorly characterized (Alfano et al., 2016b). Moreover, our data add yet another protein to the long list of FBW7 targets. Interestingly, FBW7 is among the most frequently mutated genes in cancer and therefore it is being considered as a rational target for therapeutic approaches (Welcker et al., 2008). Deregulation of FBW7 affects, as a consequence, a myriad of genes, given that many of its targets are transcription factors impacting on a wide range of players involved in many cellular pathways (Welcker et al., 2008). Similarly, also NONO with its role in mRNA regulation and in genomic stability could account at least for some of the pleiotropic effects triggered by FBW7 modulation, suggesting that the evaluation of NONO status in tumors bearing alterations of the FBW7 pathway could be crucial.
ACKNOWLEDGMENTS
The authors wish to thank the Sbarro Health Research Organization (http://www.shro.org), the Commonwealth of Pennsylvania, and the Associazione Italiana per la Ricerca sul Cancro (AIRC IG 15690) for their support. A. Giordano is also the Coordinator of the Cell Cycle and Cancer Lab at CROM, Istituto Nazionale Tumori 'Fondazione G. Pascale'-IRCCS, Naples, Italy. F. Pentimalli is also Adjunct Associate Professor at Temple University, Department of Biology, Philadelphia, PA, USA.
CONFLICTS OF INTEREST
The authors declare no conflict of interest.




