Crosstalk of ER stress-mediated autophagy and ER-phagy: Involvement of UPR and the core autophagy machinery
Abstract
Endoplasmic reticulum (ER) stress, a common cellular stress response, is closely related to the activation of autophagy that is an important and evolutionarily conserved mechanism for maintaining cellular homeostasis. Autophagy induced by ER stress mainly includes the ER stress-mediated autophagy and ER-phagy. The ER stress-mediated autophagy is characterized by the generation of autophagosomes that include worn-out proteins, protein aggregates, and damaged organelles. While the autophagosomes of ER-phagy selectively include ER membranes, and the double membranes also derive, at least in part, from the ER. The signaling pathways of IRE1α, PERK, ATF6, and Ca2+ are necessary for the activation of ER stress-mediated autophagy, while the receptor-mediated selective ER-phagy degrades the ER is Atg40/FAM134B. The ER stress-mediated autophagy and ER-phagy not only have differences, but also have connections. The activation of ER-phagy requires the core autophagy machinery, and the ER-phagy may be a branch of ER stress-mediated autophagy that selectively targets the ER. However, the determined factors that control the changeover switch between ER stress-mediated autophagy and ER-phagy are largely obscure, which may be associated with the type of cells and the extent of stimulation. This review summarized the crosstalk between ER stress-mediated autophagy and ER-phagy and their signaling networks. Additionally, we discussed the possible factors that influence the type of autophagy induced by ER stress.
1 INTRODUCTION
The endoplasmic reticulum (ER) is a dynamic organelle whose functions include protein folding, Ca2+ buffering, and lipid and carbohydrate metabolism. Diverse cellular stresses such as perturbations in Ca2+ homeostasis, redox imbalance and protein folding defects cause unfolded or misfolded proteins to accumulate in the ER lumen, a condition known as ER stress (Zhang, Wang, Chen, & Liu, 2014; Zhang et al., 2017; Zhong et al., 2011). To buffer ER stress and orchestrate the recovery of ER function, cells have an integrated signaling system, including the unfolded protein response (UPR) and ER-associated degradation (ERAD) which mainly consists of mechanisms: ubiquitin-proteasome-dependent ERAD and autophagy-lysosome dependent ERAD (Fujita et al., 2007; Schröder & Kaufman, 2005).
Autophagy is a cellular degradation process initiated in response to stress, which attempts to restore metabolic homeostasis through the catabolic lysis of aggregated proteins, unfolded/misfolded proteins or damaged subcellular organelles (Mizushima, 2007). More and more studies have shown that ER stress initiates autophagy (Bachar-Wikstrom, Wikstrom, Kaiser, Cerasi, & Leibowitz, 2013; Ogata et al., 2006; Rzymski et al., 2010). The autophagy induced by ER stress, mentioned in the previous studies, mainly includes ER stress-mediated autophagy and ER-phagy. The role and molecular mechanisms of ER stress-mediated autophagy were discussed in different conditions, such as hypoxia, ischemia/reperfusion, tumor, and so on (Chandrika et al., 2015; Ma et al., 2014; Rouschop et al., 2010). In current studies, the researchers demonstrated the activation of ER stress and autophagy by detecting expression of marker proteins. And then, they inhibited ER stress and detected the expression of autophagy, and found that the level of autophagy was reduced, which suggested that the activation of autophagy was associated with ER stress, so called it for ER stress-mediated autophagy. The ER-phagy, a selective autophagy that degrades ER, was rarely mentioned in these studies of ER stress and autophagy. The relationship between ER stress-mediated autophagy and ER-phagy is not yet fully understood. This review provided a comprehensive overview of ER stress-mediated autophagy and ER-phagy, and discussed the determined factors that control the changeover switch between ER stress-mediated autophagy and ER-phagy.
2 TYPE OF AUTOPHAGY INDUCED BY ER STRESS
2.1 ER stress-mediated autophagy
As is known to all, autophagy is activated under ER stress condition. However, the type of autophagy induced by ER stress is obscure in many studies. In the most of studies, the autophagy that is activated under ER stress condition is usually named as “ER stress-mediated autophagy.”
Ding et al. (2007) demonstrated that the activation of autophagy was depended on the induction of ER stress. They induced ER stress by treatment of murine embryonic fibroblasts with A23187, tunicamycin and thapsigargin for 24 hr. These ER stressors induced autophagy, as evidenced not only by increase of GFP-LC3 punctation (microtubule-associated protein light chain 3) and LC3-II accumulation, but also by increase of autophagic vacuoles. In addition, they also found that both double-membrane structures contain different intracellular contents in prostate cancer cell by electron microscopy. According to the above results, they thought that the ER stress-mediated autophagy was macroautophagy. However, in most studies about autophagy and ER stress, the type of autophagy induced by ER stress is obscure. Feng et al. (2017) established the mouse model of the transient middle cerebral artery occlusion (MCAO) and observed that the expression of ER stress markers increased, along with the increased of autophagy markers (ratio of LC3II/I and Beclin-1), which represented the activation of ER stress and autophagy. Moreover, ischemia/reperfusion-induced autophagy was significantly blocked by ER stress inhibitor (4-phenyl butyric acid), as well as ER related signaling inhibitors (GSK2656157 and 3, 5-dibromosalicylaldehyde). These results further demonstrated that the activation of autophagy was dependent on ER stress. Therefore, they called this process as “ER stress-dependent autophagy.” Similarly, Hart et al. (2012) found that c-Myc activated the arm of the UPR and autophagy, and inhibition of protein kinase RNA (PKR)-like ER kinase (PERK) significantly reduced c-Myc-induced autophagy in human lymphoma cells. But the contents of autophagy vacuoles were undefined.
As mentioned above, most studies demonstrated the activation of autophagy induced by ER stress by detecting the expression of autophagy markers, such as LC3II, Beclin-1, and P62 and so on. However, few studies could detect the contents of autophagic vacuoles which help to determine the type of autophagy induced by ER stress.
2.2 ER-phagy
Some studies have discovered that the ER-phagy, a new branch of macroautophagy, involves the generation of autophagosomes that selectively include ER membranes and whose delimiting double membranes also derive, at least in part, from the ER, which are different from the above studies.
To characterize the UPR-induced remodeling of the ER, Bernales, McDonald, and Walter (2006) collected exponentially growing wild-type cells treated with dithiothreitol to induce the UPR, and then carried out an ultrastructural analysis of the ER using electron microscopy. They found that folding stress increased the volume of the yeast ER significantly. Unexpectedly, in many cells ER expansion was followed by the appearance of large vesicles of 300–700 nm in diameter, which were bounded by double membranes. These vesicles were densely filled with stacked membrane cisternae, and they contained little cytosol and no other organelles, which indicated that membranes were packaged into them selectively. To determine if these membranes are derived from the ER, they examined flash-frozen/freeze-substituted sections stained with osmium, and found the delimiting outer membranes were densely studded with ribosomes, suggesting that these membranes are indeed derived from ER. They therefore referred to these vesicles as ER-containing autophagosomes, or ERAs. To obtain further evidence that the membrane stacks observed in ERAs in the electron microscopy images are indeed derived from the ER, they prepared electron microscopy images for staining with immunogold labeling of ER marker protein, and observed selective labeling of clearly identifiable ER structures, as well as selective labeling of ERAs. Hence, both the sequestered and the sequestering membranes come from the ER, indicating that the formation of ERAs involves an engulfment of the ER by itself, and this process is termed “ER-phagy.”
In short, the autophagy induced by ER stress mainly includes the ER stress-mediated autophagy and ER-phagy. The autophagosomes of ER stress-mediated autophagy include worn-out proteins, protein aggregates, and damaged organelles, while the autophagosomes of ER-phagy selectively include ER membranes, and the double membranes also derive, at least in part, from the ER. To further understand the autophagy induced by ER stress, we discussed the possible mechanisms of ER stress-mediated autophagy and ER-phagy.
3 POSSIBLE MECHANISMS OF ER STRESS-MEDIATED AUTOPHAGY
3.1 UPR and autophagy
Upon ER stress, cells activate a series of complementary adaptive mechanisms to cope with protein-folding alterations, which are known as the UPR. The UPR is a highly conserved mechanism that enables the cell to respond and to resolve the ER stress (Hetz, 2012; Patil & Walter, 2001; Ron & Walter, 2007; Rutkowski & Hegde, 2010). The UPR is characterized by three major response arms. Specifically, ER stress triggers the activity of three ER membrane resident proteins that sense the presence of unfolded proteins in the ER lumen: inositol-requiring protein-1α (IRE1α), PERK, and activating transcription factor 6 (ATF6). Under physiological conditions, these transmembrane proteins bind to the glucose-regulated protein 78 kDa (Grp78; also known as BiP) in the ER lumen. When unfolded proteins accumulate in the ER, Grp78/BiP is released from these complexes to assist with the folding of accumulated proteins, and later IRE1α, PERK, and ATF6 activate respective transducers (Figure 1).
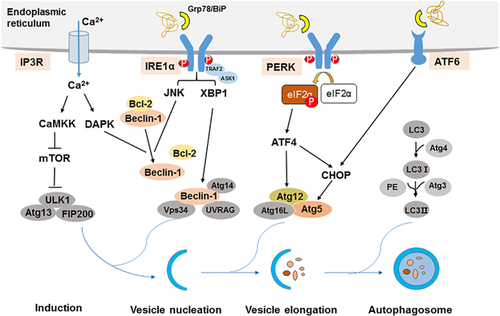
The three canonical branches of the UPR regulate autophagy at different stages in the process, induction, vesicle nucleation, and elongation. Accumulation of unfolded proteins in the ER stimulates IRE1α oligomerization in ER membranes and autophosphorylation of IRE1α's cytosolic domain. IRE1α forms a complex with tumor necrosis factor receptor-associated factor-2 (TRAF2) and apoptosis signal-regulating kinase-1 (ASK1), which causes activation downstream of stress kinases Jun-N-terminal kinase (JNK) that promote autophagy (Gardner & Walter, 2011; Ron & Hubbard, 2008). The activation JNK-mediated phosphorylation of Bcl-2 results in the disruption of the Beclin-1/Bcl-2 complex, thus releasing free Beclin-1 to form of the vacuolar protein sorting 34 (Vps34)–Beclin-1 complex, which drives the nucleation of the isolation membrane (Pattingre et al., 2005; Yamamoto, Ichijo, & Korsmeyer, 1999). In addition, X-box binding protein-1 (XBP-1), the transcription factor which is mediated by IRE1α RNAse domain, also triggers autophagy through transcriptional activation of Beclin-1 (Hetz et al., 2009; Margariti et al., 2013).
The activation of PERK inhibits general protein translation by eIF2α phosphorylation, enabling dedicated translation of transcripts harboring an alternate open reading frame, including ATF4, a key transducer (Hetz, 2012; Rutkowski & Hegde, 2010). ATF4 transcriptionally regulates Atg12, and ATF4-mediated C/EBP-homologous protein (CHOP) activation also transcriptionally induces Atg5. Atg5, Atg12, and Atg16L form Atg5-Atg12- Atg16L complex, which involves the elongation process (B'chir et al., 2013; Kang et al., 2017).
ATF6 is a transcriptional factor that upon ER stress translocates to the Golgi compartment where it is cleaved by Site-1 protease (S1P) and Site-2 protease (S2P). The cleaved N-terminal cytosolic domain of ATF6 then translocates into the nucleus where it binds to ATF/cAMP response elements Sano and ER stress-response elements to activate target genes including XBP-1 and CHOP (Adachi et al., 2008; Guo et al., 2014; Hirsch, Weiwad, Prell, & Ferrari, 2014). And then, ATF6 indirectly regulate autophagy via XBP-1 and CHOP.
3.2 Ca2+ and autophagy
ER contains a high concentration of Ca2+ and is the only cellular organelle that plays an essential role in intracellular Ca2+ homeostasis. The escalation of intracellular Ca2+ release into the cytoplasm under ER stress condition, which also involves the activation of autophagy. Ca2+ release from ER lumen through the IP3R activates a CamKK/AMPK-dependent pathway that relieves mTOR inhibition on the ULK1 complex, which plays an important role on the induction of autophagy (Borodkina et al., 2016; Høyer-Hansen et al., 2007). Death-associated protein kinase (DAPK) activation by calcium release participates in Beclin-1 phosphorylation and subsequent promotes Beclin-1 dissociation from Bcl-2, and thus induces autophagy (Simon et al., 2016; Zalckvar, Berissi, Eisenstein, & Kimchi, 2009).
In conclusion, the possible mechanisms of ER stress-mediated autophagy are complex, mainly include IRE1α, PERK, ATF6, and Ca2+ signal pathway, which regulate autophagy at different stages in the process, induction, vesicle nucleation, and elongation.
4 POSSIBLE MECHANISMS OF ER-PHAGY
To characterize the UPR-induced remodeling of the ER, Bernales, Schuck, and Walter (2007) collected exponentially growing wild-type cells treated with dithiothreitol to induce the UPR, and they found that the UPR is associated with ER-phagy, but the potential molecular mechanisms are still unclear, and need further study.
In selective types of autophagy, receptor proteins play central role in target selection. These proteins bind or localize to specific targets, and also interact with Atg8/LC3 homologues on forming autophagosomal membranes, leading to the efficient sequestration of the targets by the membranes (Okamoto, 2014). Recent studies have found that the receptor which mediate selective autophagy degrades the ER (Figure 2).
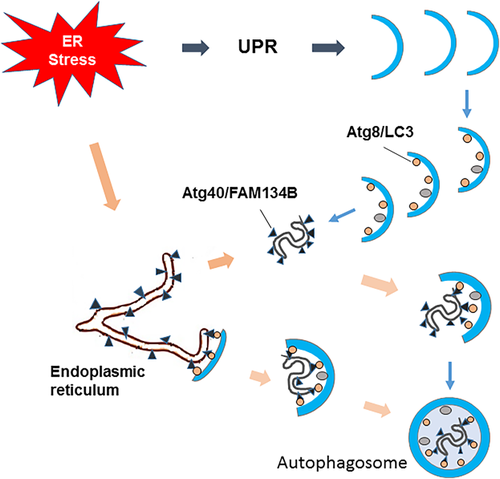
To understand comprehensively the physiological roles of selective autophagy, Mochida et al. (2015) sought to identify Atg8-binding proteins in Saccharomyces cerevisiae, which should include unknown autophagic receptors. Among proteins detected by mass spectrometry of Atg8 immunoprecipitates, they focused on two uncharacterized proteins, Ylr312c and Yor152c, which were named Atg39 and Atg40, respectively. Atg39 specifically localized to the perinuclear ER, a large proportion of Atg40 resided in the cortical ER and cytoplasmic ER, although weak localization to the pnER was also observed by fluorescence microscopy. Yeast two-hybrid assays and immunoprecipitation analysis confirmed the interactions of Atg39 and Atg40 with Atg8, and these interactions were weakened by mutations in the Atg8 pocket that binds to the Atg8-family-interacting motif. Furthermore, the protein levels of Atg39 and Atg40 markedly increased when S. cerevisiae were treated with rapamycin, and ER-phagy was blocked when knockout of either ATG39 or ATG40. All of these results demonstrated that Atg39 and Ath40 are the receptors that mediated selective autophagy degrades the ER.
Meanwhile, Khaminets et al. (2015) also found the underlying regulatory mechanisms of ER-phagy in mammalian cell. To identify proteins involved in selective autophagy, they performed yeast two-hybrid screens using LC3B and GATE16/GABARAPL2 (c-aminobutyric acid receptor-associated protein-like 2) as baits, and found that members of the reticulon-homology-domain-containing FAM134 protein family interact with LC3B and GABARAPL2, and this interaction was confirmed using FAM134B as the bait. What is more, overexpressed and endogenous FAM134B directly interacts with LC3/GABARAP in mammalian cells by Co-immunoprecipitation and GST pull-down. It is worth attention that they found that FAM134 proteins contain a conserved putative LC3-interacting region (LIR motif), and substitution of amino acids of this LIR motif with alanines (DDFELL/AAAAAA; mutLIR) completely abolished the interaction of FAM134B with LC3-like modifiers. Therefore, these results demonstrated that FAM134B was ER-resident receptors that binds to autophagy modifiers LC3 and GABARAP, which facilitates ER degradation by ER-phagy.
In conclusion, Atg40 and FAM134B, the functional counterpart, are the receptor of ER-phagy that selectively degrades the ER.
5 CROSSTALK OF ER STRESS-MEDIATED AUTOPHAGY AND ER-PHAGY
5.1 Core autophagy machinery: the necessity of ER-phagy activation
One characteristic of autophagy is the generation of autophagosomes, which are large double-membrane vesicles that engulf their cargo as they form. Some autophagy-related proteins (Atg1–Atg10, Atg12–Atg14, Atg16, and Atg18), the well conserved core autophagy machinery, is essential for the assembly of autophagosome. The remaining Atg proteins function as accessory factors and determine whether cargo engulfment is non-selective or selective (Mizushima, Yoshimori, & Ohsumi, 2011; Xie & Klionsky, 2007).
Bernales et al. (2006) treated yeast cells with dithiothreitol to induce the UPR, and they discovered that in many cells ER expansion is accompanied by the formation of autophagosome-like structures that are densely and selectively packed with membrane stacks derived from the UPR-expanded ER, and therefore named these structures as ERAs. The discovery of ERAs led them to explore the role autophagy might have in the UPR. They detected the expression of autophagy marker Atg8 with Western blotting, and found that UPR induced the expression of Atg8. Furthermore, they found that the ERAs formation were prevented after deletion of Atg8. These results indicated that the general autophagy machinery is used to generate ERAs. To identify the receptor that mediated selective autophagy degrades ER, Nakatogawa and Mochida (2015) confirmed the interactions of Atg39 and Atg40 with Atg8 by Yeast two-hybrid assays and immunoprecipitation analysis, and these interactions were weakened by mutations in the Atg8 pocket that binds to the Atg8-family-interacting motif, and thus caused partial defects in ER-phagy. Meanwhile, Khaminets et al. (2015) also found the interaction of FAM134B with LC3B in mammalian cell, which was the key factors that mediated selective autophagy degrades ER.
In short, all of studies mentioned above suggested that the core autophagy machineries involve in the ER-phagy, and are the necessity of ER-phagy activation (Figure 3).
5.2 ER-phagy: a branch of ER stress-mediated autophagy
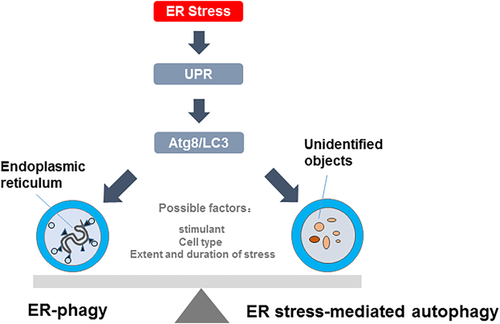
As mentioned above, both ER-phagy and ER stress-mediated autophagy are activated under ER stress condition. However, the type of ER stress-mediated autophagy is unclear, and whether ER-phagy belongs to ER stress-mediated autophagy is also unclear.
In current studies about ER stress and autophagy, the activation of ER stress and autophagy were demonstrated by detecting expression of marker proteins respectively. Furthermore, inhibition of ER stress caused partial defects in autophagy, which suggested that the activation of autophagy was associated with ER stress, and thus the autophagy was called ER stress-mediated autophagy (Periyasamy, Guo, & Buch, 2016; Tan, Muhammad, & Tan, 2016; Yu et al., 2017). Ding et al. (2007) demonstrated that the activation of autophagy was depended on the induction of ER stress, and they found that both double-membrane structures contain different intracellular contents, therefore, they thought that the ER stress-induced autophagy was macroautophagy. However, few studies could detect the contents of autophagic vacuoles which help to determine the type of autophagy induced by ER stress. When the autophagic vacuoles selectively include ER membranes, the ER stress-mediated autophagy may be ER-phagy. In addition, there are common upstream signal pathway (UPR) and core autophagy machinery between ER stress-mediated autophagy and ER-phagy. In view of the above-mentioned facts, we think that the ER-phagy may be a branch of ER stress-mediated autophagy.
5.3 Determined factors that control the changeover switch between ER stress-mediated autophagy and ER-phagy
It has been known that the ER stress can cause autophagy, including ER stress-induced autophagy and ER-phagy. However, the factors that affect the type of autophagy induced by ER are still unclear.
Xi, Barredo, Merchan, and Lampidis (2013) treated human pancreatic tumor cell with glucose starvation and observed that the expression of ER stress markers (Grp78) increased, along with the increased of autophagy markers (ratio of LC3II/I), which represented the activation of ER stress and autophagy. Moreover, starvation-induced autophagy was significantly blocked by ER stress inhibitor (4-phenyl butyric acid). These results further demonstrated that starvation contributes to the activation of ER stress-mediated autophagy. Noteworthily, Mochida et al. (2015) also established nitrogen starvation conditions, but in yeast cells, and they found that starvation induced ER-phagy, which selectively sequesters and degrades ER. The condition that induce the ER stress is similar, which both are starvation condition, but the type of autophagy is different. The difference may be due to the different cells.
Yorimitsu, Nair, Yang, and Klionsky (2006) used the drugs dithiothreitol (3 mM) and tunicamycin (2 µg/ml), which are widely used as inhibitors of disulfide bond formation and glycosylation, respectively, to induce ER stress in yeast. They then monitored autophagic induction following treatment of cells with either drug through GFP-Atg8 processing, and found that treatment with dithiothreitol or tunicamycin results in elevated levels of free GFP derived from GFP-Atg8, indicating induction of autophagy. Bernales et al. (2006) also collected exponentially growing wild-type yeast cells treated with dithiothreitol (8 mM) and tunicamycin (2 µg/ml) to induce the UPR, and they found that ER expansion was accompanied by the formation of autophagosome-like structures that are densely and selectively packed with membrane stacks derived from the ER. Moreover, they detected the expression of autophagy marker Atg8 with Western blotting, and found that UPR induced the expression of Atg8. Taken together, these results indicated that UPR induced by dithiothreitol (8 mM) and tunicamycin (2 µg/ml) contributed to ER-phagy. It is worth attention that the dithiothreitol and tunicamycin can induce autophagy, but the type of autophagy is different, which may be associated with the different extent of stimulation.
In conclusion, the activation of ER stress-mediated autophagy and ER-phagy have been found in many common conditions. The factors that control the changeover switch between ER stress-mediated autophagy and ER-phagy may be the type of cells and the extent of stimulation.
6 CONCLUSION AND PERSPECTIVES
In conclusion, ER stress-mediated autophagy and ER-phagy are closely related, but they also have differences. The activation of ER stress-mediated autophagy and ER-phagy both need the UPR and the core autophagy machinery. The difference is that ER-phagy activation need specific receptors that mediate autophagosomes formation which selective target ER, and Atg40 and FAM134B are the confirmed receptors of ER-phagy in yeast and eukaryon respectively. By contrast, it is uncertain that the autophagosomes of ER stress-mediated autophagy selectively include ER membranes, and Atg40 and FAM134B are not necessarily receptors mediated autophagy induced by ER stress. It is worth noting that the ER stress-mediated autophagy and ER-phagy may be both activated under ER stress condition, and the activatory degree may be associated with the type of cells and the extent of stimulation. However, the determined factors that control the changeover switch between ER stress-mediated autophagy and ER-phagy need much more work to be done to better understand the autophagy induced by ER stress.
ACKNOWLEDGMENTS
This work was supported by National Natural Science Foundation of China (Grant No. 81670086), National Natural Science Foundation of China (Grant No. 81370183), Tianjin Natural Science Foundation (Grant No. 14JCYBJC27800), National Training Program of Innovation and Entrepreneurship for Undergraduates (Grant No. 201510062005), and International S&T Cooperation Program of China (ISTCP, 2015DFA50310).
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.




