Identification of mitochondrial function-associated lncRNAs in septic mice myocardium
Abstract
The present study aimed to analyze long noncoding RNA (lncRNA) and messenger RNA (mRNA) expression profiles in septic mice heart and to identify potential lncRNAs and mRNAs that be responsible for cardiac mitochondrial dysfunction during sepsis. Mice were treated with 10 mg/kg of lipopolysaccharides to induce sepsis. LncRNAs and mRNAs expression were evaluated by using lncRNA and mRNA microarray or real-time polymerase chain reaction technique. LncRNA-mRNA coexpression network assay, Gene Ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed. The results showed that 1275 lncRNAs were differentially expressed in septic myocardium compared with those in the control group. A total of 2769 mRNAs were dysregulated in septic mice heart, most of which are mainly related to the process of inflammation, mitochondrial metabolism, oxidative stress, and apoptosis. Coexpression network analysis showed that 14 lncRNAs were highly correlated with 11 mitochondria-related differentially expressed mRNA. Among all lncRNAs and their cis-acting mRNAs, 41 lncRNAs-mRNA pairs (such as NONMMUG004378 and Apaf1 gene) were enriched in GO terms and KEGG pathways. In summary, we gained some specific lncRNAs and their potential target mRNAs that might be involved in mitochondrial dysfunction in septic myocardium. These findings provide a panoramic view of lncRNA and might allow developing new treatment strategies for sepsis.
1 INTRODUCTION
Sepsis, which can cause multiple organ dysfunction syndrome and shock, is an emergent medical condition with morbidity of over 19 million individuals per year.1 Recently, the mortality of sepsis has declined due to the development of effective therapeutic strategies, but septic patients with cardiac dysfunction are prone to have a poor prognosis and high risk of death.2 The pathogenesis of septic cardiomyopathy might be involved in the inflammatory cascade, activation of nitric oxide synthase, metabolic remodeling and oxidative stress.3-5 Mitochondrial dysfunction is the key event of myocardium damage during sepsis.6, 7
Long noncoding RNAs (lncRNAs) participate in many cellular biological processes.8, 9 In the past few years, numerous studies have tried to reveal specific functions of lncRNAs and possible underlying mechanisms in many pathophysiological conditions.9, 10 More and more evidence has shown that differentially expressed lncRNAs were important regulators in many heart diseases including ventricular hypertrophy, myocardial infarction, and arrhythmia.11-14 For instance, upregulated lncRNA MALAT1 in mice heart was responsible for cecal ligation and puncture-induced sepsis and myocardiac dysfunction via p38 MAPK/NF-κB signaling pathways.15 MALAT1 was also reported as a sponge for miR-320 and could induce myocardial apoptosis and increase infarct size in mice acute ischemia model through enhancing PTEN expression.16 Abnormal expression of lncRNA RMRP and NEAT1 have also been reported to be responsible for myocardial injury in sepsis models.17, 18 Mitochondrial function can be regulated by lncRNAs in cancer, which might act as a valuable target for a new strategy of cancer treatment.19 Whether the mitochondrial dysfunction is modulated by lncRNAs in septic cardiomyopathy and whether other lncRNAs also take part in the pathophysiology of sepsis remain unknown.
The objective of this research was to analyze lncRNA and messenger RNA (mRNA) expression profiles in mice septic cardiac injury model and to identify the potential lncRNAs and mRNAs that might be responsible for cardiac mitochondrial dysfunction during sepsis.
2 MATERIALS AND METHODS
2.1 Reagents
TAKARA RNAiso Plus was from Takara Bio (Mountain View, CA). NucleoSpin RNA clean-up XS kit was from Macherey-Nagel (Düren, Germany). RNeasy mini kit and RNase-free DNase set were obtained from QIAGEN (Hilden, Germany). ReverTra Ace qPCR kit was from Toyobo (Tokyo, Japan). Low Input Quick Amp Labeling kit and Gene Expression Hybridization kit were obtained from Agilent Technologies (Santa Clara, CA). Assay kits for ATP, superoxide dismutase (SOD), glutathione peroxidase (GSH-Px), and malondialdehyde (MDA) were purchased from Beyotime Institute of Biotechnology (Haimen, Jiangsu, China). Lipopolysaccharides (LPS) was from Merck KGaA (Darmstadt, Germany).
2.2 Animal model
Adult male C57B6/L mice (7-8 week and 20-25 g) were purchased from Zhejiang University Laboratory Animal Research Center. Twenty-eight mice were randomly divided into two groups receiving an intraperitoneal injection of 10 mg/kg LPS or saline solution as a control. Six hours after injection, all mice were killed. Ten cardiac tissue samples from each group were used to evaluate the mitochondrial function and morphology. Four samples from each group were immediately removed and frozen in liquid nitrogen for the detection of expression of lncRNAs and mRNAs. All animal experiments were approved by the Experimental Animal Ethics Committee of Zhejiang University. A double-blind method was used in this study.
2.3 Assessment of mitochondrial function
Since mitochondria are the main source of intracellular free radical species and energy, levels of oxidative stress and ATP in mice hearts were measured to evaluate the mitochondrial function. Heart samples were homogenized and lysed in lysis buffer. SOD activity, GSH-Px activity, and MDA (an index of lipid peroxidation) content were measured by using commercial assay kits. Protein content in the samples was evaluated using a bicinchoninic acid assay method. All data were normalized with the protein concentration of the samples. ATP level of whole tissue was measured by using the ATP Assay kit. The concentration of ATP was expressed as OD370 and normalized with protein concentrations of each sample.
2.4 Evaluation of mitochondrial morphology
Left ventricle samples were fixed in 2.5% glutaraldehyde and 1% OsO4 for 1.5 hours, then dehydrated with a series of graded alcohol. After that, samples were embedded and sliced at 50 nm, stained with 5% uranyl acetate followed by a lead citrate solution. Slices were then observed under a transmission electron microscope. Mitochondrial morphology evaluation was performed according to the Flameng mitochondrial score.20
2.5 RNA extraction
Total RNA was isolated from mice left ventricles using TAKARA RNAiso Plus following the manufacturer's instructions. After RNA integrity was checked, total RNA was further purified by NucleoSpin RNA Clean-up XS kit and RNase-Free DNase Set.
2.6 LncRNA and mRNA microarray
Total RNA was amplified and labeled with Cy3 by using Low Input Quick Amp Labeling kit. Cy3-labeled cRNA was then purified by using RNeasy mini kit.
LncRNA array or mRNA array slides were hybridized with Cy3-labeled cRNA for 17 hours. After washed with Wash Buffer, lncRNA array slides and mRNA array slides were scanned by Agilent Microarray Scanner. Raw data were normalized by Quantile algorithm, limma package the R program.
2.7 Construction of lncRNA-mRNA coexpression network
The interaction between the top 10 differentially expressed lncRNAs and all mRNAs was analyzed by calculating Pearson's correlation coefficients based on the lncRNAs and mRNAs expression levels of samples. The lncRNA-mRNA pairs with Pearson's correlation coefficient more than .99 and P < .05 were identified and visualized using Cytoscape Consortium V_3.7.1 software (San Diego, CA).
2.8 Prediction of lncRNA targeted genes
To predict the cis-regulatory target of lncRNAs, we selected the intergenic lncRNAs and searched the protein-coding locus nearby (within <10 kb). These protein-coding genes are considered to be the cis-regulatory targets of the lncRNA. For the trans-regulatory target prediction, the mRNA sequences which are complementary to lncRNAs were searched by using BLAST software. Then the complementary energy between every pair of lncRNA-mRNA sequences was calculated by using RNAplex program. The genes with complementary energy less than or equal to −30 were identified as the possible trans-regulatory targets of lncRNAs.
2.9 Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analysis
Functions in biological pathways or Gene Ontology (GO) terms of lncRNAs were analyzed through their predicted targeting mRNAs according to the latest GO database (http://www.geneontology.org) and Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/). The GO analysis and KEGG pathway analysis were performed using the “clusterProfiler” package in R software with Bioconductor annotation package “org.Mm.eg.db”. A two-sided Fisher's exact test was used and P < .05 was set as the threshold value. Then the top 30 enriched terms and KEGG pathways were visualized by dotplot. The same analysis was also performed on the differentially expressed mRNAs.
2.10 Quantitative real-time polymerase chain reaction
Total RNA was reverse-transcribed and then complementary DNA was synthesized by using ReverTra Ace qPCR kit. Quantitative real-time polymerase chain reaction (qRT-PCR) was conducted by a QuantStudio 5 Real-Time PCR System (ABI, Foster City, CA). The specific primer pairs were shown in Table S1. Glyceraldehyde 3-phosphate dehydrogenase mRNA was used as a housekeeping gene to normalize mRNA levels between different samples. Relative gene expression was determined by using 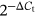 method or fold change in expression.
method or fold change in expression.
2.11 Statistical analysis
Data were expressed as mean ± standard error of mean. Student's t test was used for analysis (GraphPad Prism 8.0).
3 RESULTS
3.1 Evaluation of mitochondrial function and morphology of septic mice hearts
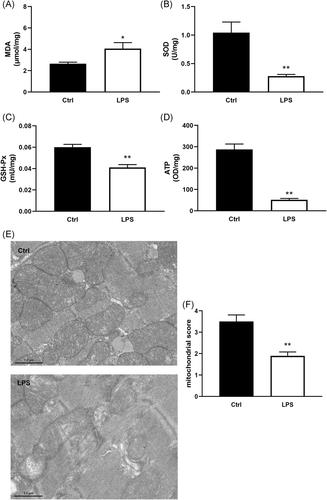
Mitochondrial damage is believed to be a key factor leading to a worse prognosis during septic cardiomyopathy. The results showed that MDA content was increased, while SOD and GSH-Px activities were decreased in LPS-treated mice heart (Figure 1A-C). Consistent with these results, the ATP level in cardiac tissue was also lower in LPS-induced septic mice than that of control (Figure 1D). Transmission electron microscope analysis revealed an obvious swelling and loss of cristae of mitochondria in LPS-treated mice ventricular tissue when compared with the control group (Figure 1E,F).
3.2 LncRNA and mRNA expression profiles in septic mice heart
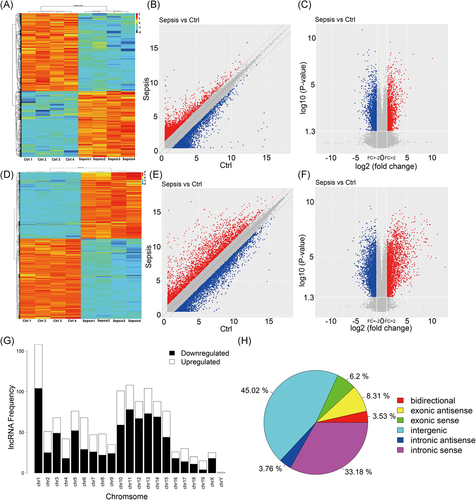
Microarray analysis was performed on four heart samples from the control group and four from the LPS group to detect the differential expression of lncRNAs and mRNAs. Compared with control group, 471 upregulated lncRNAs and 804 downregulated lncRNAs with a change of more than twofold (P < .05) were identified in the LPS group. We also found 1158 upregulated mRNAs and 1611 downregulated mRNAs in the LPS group when compared with that of the control group (Figure 2A-F).
3.3 Expression characteristics of dysregulated lncRNAs in septic mice heart
The genomic locations of the 1275 dysregulated lncRNAs are shown in Figure 2G, according to their neighboring protein-coding genes. These differentially expressed lncRNAs can be divided into six groups (Figure 2H). Intergenic RNA and intronic sense RNA account for a large proportion (45.02% and 33.18%, respectively). Exonic antisense and exonic senses are 8.31% and 6.2%, respectively. The remaining two are bidirectional and intronic antisense.
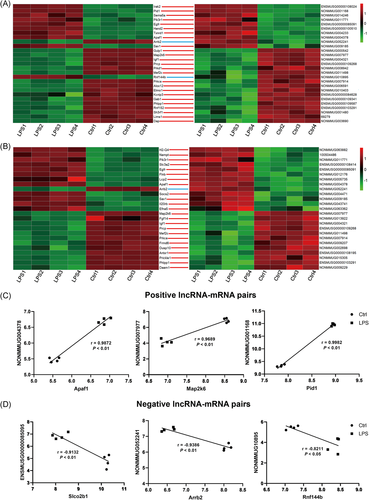
3.4 Construction of lncRNAs-mRNAs coexpression network and function analysis
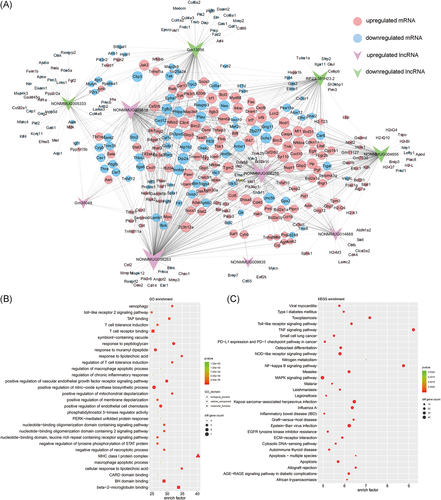
To predict the potential function of differential expressed lncRNAs, a lncRNA-mRNA coexpression network was constructed according to top 10 differentially expressed lncRNAs (six upregulated and four downregulated). All lncRNA-mRNA pairs with high linear correlation (Pearson correlation coefficient >.99 or <−.99 for positive or negative correlation, respectively) and P < .05 were shown in Figure 3A. A total of 343 mRNAs were identified, which comprise 934 specific lncRNA-mRNA coexpression relationships. Among this network, most of the lncRNAs are considered to co-express with more than 10 mRNAs, while 9 mRNAs (Dip2a, Hand2, Tgm2, Antxr1, Nod1, Nfkb2, Cxcl10, Birc2, and Nfkbia) are coexpressed with the greatest number of those top 10 lncRNAs. Then the function of these 343 mRNAs was analyzed by using GO and KEGG enrichment analysis (Figure 3B,C). The results showed that these mRNAs mainly take part in inflammation-related and mitochondria-related pathways, such as Toll-like receptor (TLR) signaling, NF-κB signaling, MAPK signaling, TNF signaling, regulation of mitochondrial depolarization, and apoptosis.
3.5 Function analysis of cis- or trans-regulatory target genes of differentially expressed lncRNAs
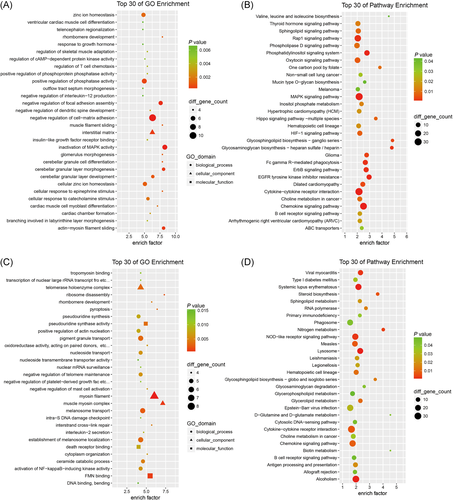
Since most lncRNAs are localized in the nucleus, the differentially expressed lncRNAs might modulate their target genes through cis- or trans-regulatory mechanisms. Among genes 10 kb up- and downstream of lncRNAs, 872 differentially expressed lncRNAs were found colocalized with 1170 cis-regulated genes. By using BLAST software and RNAplex program, 801 differentially expressed lncRNAs with their 1674 trans-regulated genes were also identified. According to these cis- or trans-regulated target genes, potential functions of these differentially expressed lncRNAs in septic mice hearts were analyzed by using GO and KEGG assay. The results (Tables S2-S5) revealed that the target genes of differentially expressed lncRNAs were highly enriched in processes like inflammation and immune activity. For lncRNA cis-target mRNAs, the most significant enriched GO term is inactivation of MAPK activity (Figure 4A). While in the KEGG pathway, the MAPK signaling pathway is also a highly enriched term (Figure 4B). For lncRNA trans-target mRNAs, similar terms which are related to inflammation, mitochondrial metabolism, and contractile function were also identified in GO terms (such as FMN binding, oxidoreductase activity, activation of NF-κB-inducing kinase activity, and myosin filament) and in KEGG pathways (such as viral myocarditis, NOD-like receptor signaling pathway) (Figure 4C,D).
3.6 Expression profiles of mRNAs related to mitochondrial dysfunction
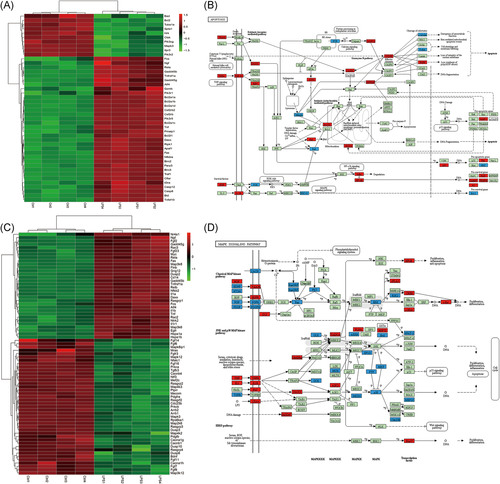
Given that mitochondrial dysfunction is a universal and critical process mediating cardiomyocyte apoptosis and inflammatory responses, expression profiles of mitochondrial function-related mRNAs in mice hearts were analyzed. A total of 384 differentially expressed mRNAs in GO terms associated with mitochondrial function (eg, GO: 0006915 apoptotic processes, GO: 0072593 regulation of reactive species metabolic process, GO: 0033198 response to ATP, and GO: 0006979 response to oxidative stress.) were detected (Figure S1 and Table S6), and 355 mRNAs in mitochondrial function-related KEGG pathways (eg, mmu04064: NF-κB signaling pathway, mmu05416: viral myocarditis, mmu04210: apoptosis, mmu04010: MAPK signaling pathway) were identified (Figure S2 and Table S7).
Dysregulated mRNAs in the apoptosis pathway and MAPK signaling pathway were further investigated. It has found that 32 mRNAs were upregulated and 11 mRNAs were downregulated in the apoptosis pathway (Figure 5A,B). The upregulated mRNAs include Fos, Casp8 and Bid, and the downregulated mRNAs include Bad and Bcl2, which indicate the activation of the mitochondrial apoptosis pathway. While in MAPK signaling pathway, dysregulated mRNAs, such as Map3, Rasgrp1and Ngf were identified (Figure 5C,D).
3.7 Coexpression network of mitochondria-related differentially expressed mRNAs and lncRNAs
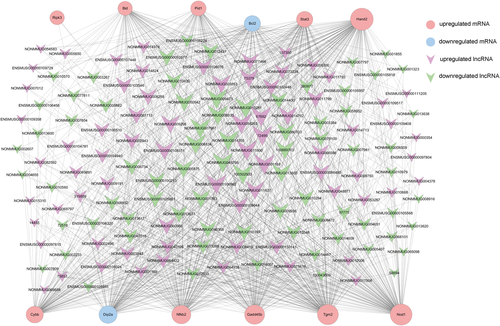
To predict the potential lncRNAs that might target mitochondria-related differentially expressed mRNA in mice heart during sepsis, we selected 12 significantly dysregulated mRNAs that play important roles in mitochondrial dysfunction, and their correlation coefficients with all 1275 lncRNAs were calculated. A total of 357 related lncRNAs with the Pearson correlation coefficient of more than .99 (P < .05) were showed in the co-expression network (Figure 6). There are 14 lncRNAs showing correlation with all of 11 mRNAs in the 12 selected mRNAs. LncRNA NONMMUG075618, one of the top 10 upregulated lncRNA in septic mice heart, had positive correlations with Gadd45b, Stat3, and other 4 upregulated mRNAs which have been proven to regulate ATP biosynthesis, oxidative stress, mitochondrial mitophagy, and mitochondrial DNA synthesis. It was also shown that NONMMUG075618 has negative correlations with Dip2a, which have protective functions on cells.
3.8 Expression profiles of lncRNAs related to mitochondrial dysfunction associated cis-regulatory target genes
Among all lncRNAs and their cis-acting mRNAs, a total of 41 lncRNAs-mRNA pairs, which might be associated with mitochondrial dysfunction, were found enriched in GO terms and KEGG pathways (Figure 7A,B). And 11 pairs were shown in both GO terms and KEGG pathways. Three positively related lncRNA-mRNA pairs and three negatively related lncRNA-mRNA pairs were selected to further analyze their correlations (Figure 7C,D). Apaf1 gene, which was previously proved to encode a cytoplasmic protein and initiate apoptosis, was increased in septic mice hearts. The expression of its cis-upstream lncRNA NONMMUG004378 was also upregulated, which was strongly correlated with Apaf1 gene with a high positive correlation coefficient (r = .9872; P < .01). The other five pairs (eg, NONMMUG007977 and Map2k6 gene, NONMMUG004378, and Pid1 gene) with high correlation coefficient are also shown in Figure 7C,D.
3.9 Validation of the microarray data by using qRT-PCR
To confirm the microarray analysis results, two lncRNAs and two mRNAs were randomly selected to validate their expression levels in two groups using the qRT-PCR assay. The data showed that lncRNA ENSMUSG00000111531 and mRNA Akap5 were downregulated, lncRNA NONMMUG075618 and mRNA Gadd45b were upregulated in LPS-treated mice hearts (Figure 8A,B), which had a high consistency with the microarray data.
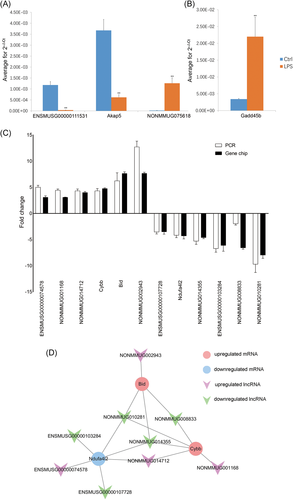
Since our particular interest lies in mitochondrial function-associated lncRNAs, three mRNAs (Cybb, Ndufa4l2, and Bid) which are responsible for ROS formation, ATP production, and mitochondrial morphology were selected. The expression of these mRNAs and nine highly associated lncRNAs were measured by using qRT-PCR. The qRT-PCR results showed that Cybb, Bid, and four lncRNAs (such as ENSMUSG00000074578 and NONMMUG014712) were upregulated, while Ndufa4l2 and other five lncRNAs (such as ENSMUSG00000107728 and NONMMUG014355) were downregulated in septic mice hearts, which were concordant with the microarray data (Figure 8C). Among these lncRNAs, NONMMUG010281 and NONMMUG014355 are highly associated with all of these three mRNAs, while NONMMUG008833 and NONMMUG014712 are highly associated with two of these mRNAs (Figure 8D).
4 DISCUSSION
Cardiovascular injury is a kind of severe complication in sepsis.1 The pathophysiological events of septic cardiomyopathy mainly include inflammation, oxidative stress, energetic deficiency, dysregulated autophagy, and apoptosis.3, 7, 21 Some lncRNAs such as MALAT1, H19, HOTAIR have been reported to be involved in regulating these activities in septic heart.15, 22, 23 In this study, a septic cardiomyopathy mice model was established to detect the differentially expressed lncRNAs and mRNAs in septic and normal mice hearts using microarrays technology. A total of 471 upregulated lncRNAs and 804 downregulated lncRNAs were identified in septic hearts. These differentially expressed lncRNAs are mainly intergenic and intronic sense. GO and KEGG pathway enrichment analysis revealed that most of them are related to immune activities (such as TLR signaling pathways, MAPK signaling pathways) and apoptosis, which is in consistence with the pathophysiological changes in septic cardiomyopathy.24-26
LncRNAs regulate gene expression in multiple ways including transcription, posttranscription, and epigenetic mechanism.27-29 Most lncRNAs were found in the nucleus. Nuclear lncRNAs have been demonstrated to regulate their neighboring genes via cis regulation, or modulate genes on other chromosomes by trans acting.30 A small number of cytoplasmic lncRNAs may act as competing endogenous RNAs to exert their regulatory effects on target genes.22 In this study, we predicted cis- or trans-regulated target genes based on dysregulated lncRNAs in septic hearts and found that they were highly enriched in inflammation, mitochondrial function, apoptosis, and contractile function.
Many studies have shown that mitochondrial function can also be regulated by lncRNAs. For instance, lncRNA MEG3 regulates mitochondrial pathway-dependent apoptosis in prostate cancer tissues.31 LncRNA SAMMSON acts as a modulator of mitochondrial homeostasis in melanoma.32 LncRNA UIHTC protected cardiomyocytes against ischemic injury through improving mitochondrial bioenergetics.33 In this study, according to mitochondria-related differentially expressed mRNAs, a total of 357 related lncRNAs and 14 highly related lncRNAs were found through establishing of co-expression network.
Among these lncRNAs, NONMMUG075618 is not only highly upregulated in septic hearts, but also has a strong correlation with multiple mitochondria-related mRNAs. Thus, we hypothesized that the upregulated lncRNA NONMMUG075618 might play a crucial role in septic cardiomyopathy. Meanwhile, a total of 41 lncRNAs-mRNA pairs with cis-acting which were associated with mitochondrial dysfunction have been identified. NONMMUG075618 was not in the list, suggesting that it might regulate the mitochondrial dysfunction-related mRNA in another manner.
Decreased ATP production, oxidative stress, and abnormal morphology of mitochondria are the typical characteristics of mitochondrial damage in septic cardiopathy. ATP production is controlled by the mitochondrial electron transport chain.34 Ndufa4l2 gene encodes Ndufa4l2 protein which is a subunit of complex I of the electron transport chain. NADPH oxidase 2 (NOX2) protein, encoded by the Cybb gene, is one of the major sources of ROS production.35 Our study showed that Ndufa4l2 was downregulated and Cybb gene was upregulated in septic mice heart. The expression of Bid gene was also found to increase in septic cardiac tissue. Bid protein, one of the proapoptotic BH3-only proteins, has been demonstrated to trigger Bax oligomerization and induce mitochondrial outer membrane permeabilization, leading to mitochondrial hyperfusion and abnormality in morphology.36 We found that NONMMUG010281 and NONMMUG014355 are all highly associated with these three genes, suggesting that they might be the main lncRNAs that regulate the cardiac mitochondrial function and morphology during sepsis.
Mitochondrial function-related lncRNAs can be mainly divided into two types: mitochondria-encoded lncRNAs (mtlncRNAs) and nuclear-transported mitochondria-associated lncRNAs (ntmtlncRNAs).19, 37 For example, abnormal expression of mtlncRNAs, such as ND5 and ND6, could damage the electron transport chain and decline mitochondrial oxidative phosphorylation.38, 39 Some ntmtlncRNAs (such as RMRP and LINC00116) could regulate mitochondrial respiration and metabolic reprogramming through human antigen R-dependent mitochondrial translocation.40-42 Although we have identified the differentially mitochondrial dysfunction-related lncRNAs in septic mice hearts, the location and mechanism of that lncRNA deserve further investigation in the future.
There were some limitations to this study. Since microarray has better sensitivity and accuracy than RNA sequencing (RNA-seq) for quantification of low abundance RNAs, microarray was chosen to analyze lncRNAs in this study. However, RNA-seq has an edge over microarray on providing more valuable information about unknown sequences.43 We only screened the known lncRNAs but might overlook some potentially unknown lncRNAs sequences and novel transcript isoforms. Next, although some differentially expressed lncRNAs and mRNAs detected in microarray have been validated by using PCR, the low throughput PCR method is difficult to analyze a whole lncRNAs expression profile. PCR results cannot be representative of thousands of dysregulated RNAs found by microarray analysis. The detailed information about every single specific lncRNA (such as expression level and target mRNAs) await further investigation. Finally, although we identified some potential mitochondrial dysfunction-related lncRNAs (such as NONMMUG075618, NONMMUG010281, and NONMMUG014355), the molecular mechanisms of these lncRNAs in septic cardiopathy need to be explored in vivo or in vitro.
5 CONCLUSION
In summary, we explored the differentially expressed lncRNA and mRNA profiles of the myocardium in septic mice hearts and proposed some specific lncRNAs and their potential target mRNAs that might lead to mitochondrial dysfunctions. These findings provide us a panoramic view of lncRNA which would likely be involved in the mitochondrial dysfunction in septic cardiomyopathy and help explore new treatment strategies for sepsis from the perspective of lncRNA.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (No. 81871541). The authors acknowledge the SHBIO Biotechnology Corporation (Shanghai, China) for experimenting with lncRNA and mRNA microarray.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Shen Y and Chen Y designed the experiment and revised the manuscript. SY and ZX performed gene differential analysis and the qRT-PCR experiment. SY performed the statistical analysis and drafted the manuscript. ZM and WL evaluated the morphology and function of mitochondria. All authors read and approved the final manuscript.
ETHICS STATEMENT
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Open Research
DATA AVAILABILITY STATEMENT
All the microarray data in this study have been deposited in the Gene Expression Omnibus (GEO) database and are accessible through the accession numbers GSE142615 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142615).