Circular RNA circ_0011269 sponges miR-122 to regulate RUNX2 expression and promotes osteoporosis progression
Abstract
Circular RNAs (circRNAs) are a novel class of noncoding RNAs that are widely expressed in human disease. However, circRNAs expression profile and potential mechanism in osteoporosis pathogenesis remain to be further studied. In the present study, a total of 69 circRNAs were identified to be abnormally expressed in osteoporosis patient samples by microarray and bioinformatics analyses. We found that circ_0011269 was notably downregulated in osteoporosis (fold change, 3.94). By means of miRanda algorithm, we constructed the interaction network of circ_0011269-miRNAs in osteoporosis based on target binding and miR-122 was enrolled in the network. Dual-luciferase reporter assay verified the target relationship of miR-122 and circ_0011269/RUNX2. The expression of circ_0011269 and RUNX2 were gradually increased during osteogenic differentiation while miR-122 exhibited a decreased expression. Moreover, overexpression of circ_0011269 could promote RUNX2 expression and inhibit osteoporosis. In summary, this study found that circ_0011269 sponges miR-122 to regulate RUNX2 expression and promotes osteoporosis progression.
1 INTRODUCTION
Osteoporosis is the most common skeletal disorder worldwide, accompanied by fragility fracture and bone mass loss.1, 2 The main cause for osteoporosis is the abnormal activation of osteoclasts.3 With the development of the medical science, the estimate, and treatment for osteoporosis have been gradually improved,4 but there are still some limitations since the pathogenesis of osteoporosis is elusive.
CircRNAs are a novel class of noncoding RNAs that widely expressed in human tissues and play important roles in disease pathogenesis.5, 6 MicroRNAs (miRNAs) are small endogenous RNAs that dysregulated in various human cancers.7, 8 It was reported that circRNAs could act as miRNAs sponges by competing with the binding sites of messenger RNAs (mRNAs), and thus altered the expression of mRNAs.9-11 For example, circular RNA circ_000203 was found to enhance the expression of fibrosis-associated genes by sponging miR-26b-5p in cardiac fibroblasts.12 However, circRNAs in osteoporosis remain largely unknown. RUNX2 belongs to the family of RUNT related transcription factor and plays vital role in the osteogenic differentiation of bone marrow mesenchymal stem cells into osteoblasts.13, 14 Bone sialoprotein (BSP), osteocalcin (OCN), osteopontin (OPN), and alkaline phosphatase (ALP) activity are representative makers of osteoblasts and are used to access osteogenic differentiation.15
In the present study, we explored circRNAs expression profiles in osteoporosis patient samples by microarray and bioinformatics analyses. Circ_0011269 was notably downregulated in osteoporosis as identified by sequencing data. Circ_0011269 was located at chromosome 1: 31791013-31811895 and back-spliced from ZCCHC17. Further quantitative reverse transcription-polymerase chain reaction (qRT-PCR) experiment confirmed that circ_0011269 was lowly expressed in osteoporosis. Subsequently, the interaction network of circ_0011269-miRNAs was constructed to explore the potential mechanism of circ_0011269. Dual-luciferase reporter assay verified the target relationship of miR-122 and circ_0011269/RUNX2. Finally, circ_0011269 promoted osteoporosis progression and regulated RUNX2 expression by sponging miR-122 was verified by functional experiments. In conclusion, this study found that circ_0011269 sponges miR-122 to regulate RUNX2 expression and promotes osteoporosis progression.
2 MATERIALS AND METHODS
2.1 Collection of samples
The clinical serum or plasma samples were collected from 21 patients (Age, 55-67 years) who were diagnosed as osteoporosis in Shandong Provincial Hospital affiliated to Shandong University and 20 healthy controls (Age, 56-69 years). All participants were free from chemotherapy and metabolic diseases (diabetes, hyperparathyroidism, and systemic lupus erythematosus, etc). This study was approved by Ethics Committee of Shandong Provincial Hospital affiliated to Shandong University and the informed consents were signed by all participants. The study was in accordance with Helsinki Declaration.
2.2 RNA extraction, microarray, and bioinformatics analyses
Total RNAs were extracted from osteoporosis samples and control samples using Trizol reagent (Invitrogen, CA) according to manufacturer's instruction. The integrity and quantity of total RNAs were evaluated and then digested by RNase R to remove linear transcripts. The microarray analysis was conducted in five osteoporosis samples and five control samples by Agilent CircRNA Array V2 following the instructions. R package limma16 was carried out to normalize the expression data and calculate the differential expression. The circRNAs with fold change >2 and P < .01 were considered to be differentially expressed. The target binding of circRNAs and miRNAs were predicted by miRanda algorithm17 and the network was drawn with Cytoscape software.18
2.3 Osteogenic differentiation and dual-luciferase reporter assay
The human bone marrow mesenchymal stem cells (hBMSCs) were purchased from Ransu biotech company (Ransu, Shanghai, China) and cultured in 24-well plates at 37℃ with minimum essential medium supplemented with FBS, penicillin, l-Glutamine, etc. The induction of hBMSCs was then performed according to the protocol. The osteogenic induction medium was supplied with osteogenic inducers and replaced every 3 days. Cells were harvested for 21 days and the analysis were conducted every 7 days (0, 7th, 14th, 21th). The dual-luciferase reporter assay was performed to verify the target binding sites. Wild-type of RNAs (WT) and mutant of RNAs (MUT) were synthesized by Biotech company (Biotech, Shanghai, China) and cloned into the pGL3 Basic luciferase vectors. The luciferase activities were measured by a microplate luminometer (Thermo, Shanghai, China) after 48 hours of incubation.
2.4 qRT-PCR and Western blot
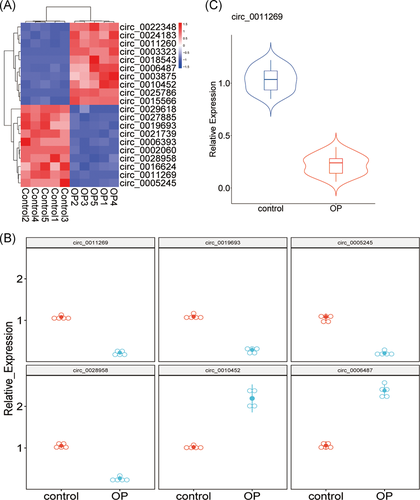
Total RNAs were transcribed to complementary DNA (cDNA) using Revert AidTM First Strand cDNA Synthesis Kit (Fermentas, Glen Burnie, MD) according to manufacturer's instruction. The qRT-PCR was performed using SYBR Green supermix (Fermentas) on a Real-time PCR Detection System. RUNX2: 5′-TGGTTACTGTCATGGCGGGTA-3′ (forward) and 5′-TCTCAGATCGTTGAACCTTGCTA-3′ (reverse); MiR-122: 5′-TCTTCCTGGAATTCAAGCCTTT-3′ (forward) and 5′-AGTGGGCCTAGTGCTGGAAA-3′ (reverse); Circ_0011269: 5′-CTAAGGAGTCACAGGAAGACATC-3′ (forward) and 5′-GTAGAATCTCTCAGACTCAAGGTTG-3′ (reverse). Relative expression levels were computed by the 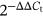 method and glyceraldehyde 3-phosphate dehydrogenase (GAPDH)/U6 were used as internal controls. Proteins were extracted from hBMSCs and then the concentration was measured. After that, proteins (10 μg) were boiled and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12%), then transferred into polyvinylidene fluoride (PVDF) membranes (Millipore, MA). The membranes were incubated with primary antibodies overnight and then re-probed with secondary antibodies. The primary antibodies for GAPDH, BSP, OCN, OPN, RUNX2 are as following: ab9485 (1:2500), ab52128 (1:500), ab93876 (1:500), ab216406 (1:1000), ab23981 (1 μg/mL). The blots were exposed using enhanced chemiluminescence following the instructions.
method and glyceraldehyde 3-phosphate dehydrogenase (GAPDH)/U6 were used as internal controls. Proteins were extracted from hBMSCs and then the concentration was measured. After that, proteins (10 μg) were boiled and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12%), then transferred into polyvinylidene fluoride (PVDF) membranes (Millipore, MA). The membranes were incubated with primary antibodies overnight and then re-probed with secondary antibodies. The primary antibodies for GAPDH, BSP, OCN, OPN, RUNX2 are as following: ab9485 (1:2500), ab52128 (1:500), ab93876 (1:500), ab216406 (1:1000), ab23981 (1 μg/mL). The blots were exposed using enhanced chemiluminescence following the instructions.
2.5 Cell transfection and ALP activity measurement
The hBMSCs were transfected with circ_0011269, miR-122 mimics and negative controls by Lipofectamine 2000 (Invitrogen) following the instruction of the manufacturer. Cells were transfected for 6 hours. The treated hBMSCs were cultured for 14 days to perform ALP staining. A commercial ALP kit was used to measure the ALP activity following the instruction of manufacturer's protocol. The absorption value at 405 nm was recorded by the spectrophotometer.
2.6 RNA immunoprecipitation
The RNA immunoprecipitation (RIP) experiment was performed in miR-122 transfected cells and control cells with Magna RIPTM Kit (Millipore) following the manufacturer's instruction. The cells (107) were lysed in RNA lysis buffer and then conjugated to the human anti-Argonaute 2 (AGO2) antibody (Millipore) or control mouse IgG (Millipore) by magnetic beads. Beads were washed with RIP buffer and the extracted RNA was detected by qRT-PCR.
3 RESULTS
3.1 Study subjects and differentially expressed cicRNAs in osteoporosis
The clinical parameters of participants enrolled in this study were listed in Table 1, including age, bone mineral density (BMD), T-score, etc. By means of microarray technology and bioinformatics analyses, 69 circRNAs were identified to be differentially expressed in osteoporosis compared to control group (n = 5, respectively), including 34 upregulated circRNAs and 35 downregulated circRNAs (data not shown). As heatmap shown in Figure 1A, top 20 abnormally expressed circRNAs with a significant differences were further screened concerning their potential function in osteoporosis. To validate the microarray data, six circRNAs were selected to measure their expression in osteoporosis samples (n = 5) using qRT-PCR, including circ_0011269, circ_0019693, circ_0005245, circ_0028958, circ_0010452, and circ_0006487. As shown in Figure 1B, the expression patterns of circRNAs were consistent with microarray data, for example, circ_0011269 was notably downregulated in osteoporosis (fold change, 3.94; P = .009), similar with microarray data (fold change, 5.98; P < .01).
| Clinical parameters (mean ± SD) | Osteoporosis | Control |
|---|---|---|
| Age | 61 ± 5.00 | 62 ± 6.00 |
| Height, cm | 169 ± 4.79 | 168 ± 5.17 |
| Weight, kg | 64 ± 8.68 | 63.8 ± 7.88 |
| BMD, g/cm2 | 0.66 ± 0.07 | 0.84 ± 0.11 |
| T-Score lumbar spine (L2-L4) | −2.7 ± 0.79 | 0.5 ± 0.86 |
| CROSSL, ng/mL | 0.78 ± 0.12 | 0.51 ± 0.06 |
| TPINP, ng/mL | 64.3 ± 8.08 | 51.9 ± 7.09 |
| OSTEOC, ng/mL | 23.5 ± 2.7 | 14.88 ± 2.6 |
- Abbreviation: BMD, bone mineral density.
3.2 Circ_0011269 was lowly expressed in osteoporosis
To further validate the expression of circ_0011269 in osteoporosis, we performed qRT-PCR in a larger osteoporosis sample size (n = 21). The result demonstrated that circ_0011269 was lowly expressed in osteoporosis samples compared to control (Figure 1C).
3.3 Circ_0011269-miRNAs interaction network
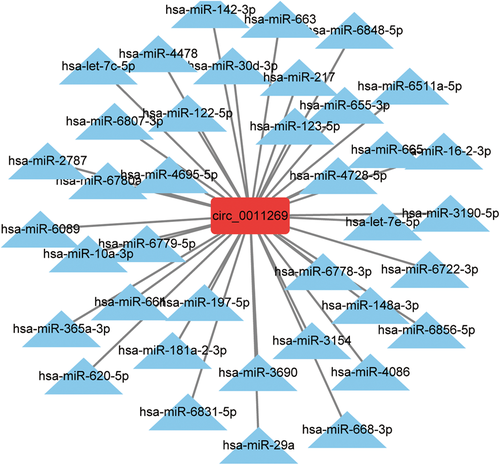
To explore the function of circ_0011269, we constructed circ_0011269-miRNAs interaction network based on the bioinformatics algorithm. In total, 38 miRNAs were indicated to have the potential binding sites of circ_0011269, as shown in Figure 2.
3.4 miR-122 contains the binding sites of circ_0011269 and RUNX2
Among the predicted miRNAs in the network, we found that miR-122 contains the binding sites of RUNX2 (Figure 3A) which predicted by miRanda algorithm. Then, the target binding of miR-122 with circ_0011269 and RUNX2 were validated using dual-luciferase reporter assay. As shown in Figure 3B,C, miR-122 inhibited the fluorescence intensity in wild-type of circ_0011269 and RUNX2, while the inhibition effect was blocked by binding motif mutation of circ_0011269 and RUNX2. Hence, the target relationship of miR-122-circ_0011269 and miR-122-RUNX2 was verified. Besides, we conducted anti-AGO2 RIP assay. The result showed that circ_0011269 pulled down with anti-AGO2 was enriched in overexpressed miR-122 cells compared with control (Figure 3D). Moreover, qRT-PCR demonstrated that the expression of miR-122 was significantly higher in osteoporosis samples (n = 5) while RUNX2 exhibited lower expression compared to control (Figure 3E).
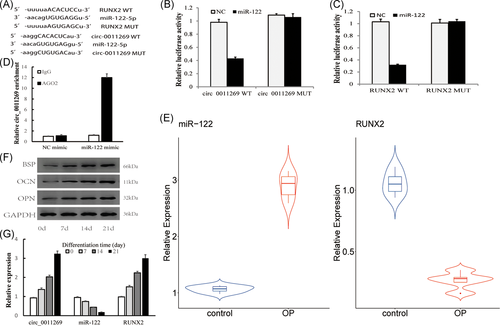
3.5 The expression of circ_0011269, miR-122, and RUNX2 during osteogenic differentiation
To explore the potential role of circ_0011269/miR-122/RUNX2 in osteoporosis, we measured the expression of circ_0011269, miR-122 and RUNX2 at 0, 7th, 14th, and 21th day during osteogenic differentiation. As shown in Figure 3F, the osteogenic-associated biomarkers (BSP, OCN, OPN) exhibited an increased expression during osteogenic differentiation. The expression of circ_0011269 and RUNX2 were gradually increased while miR-122 exhibited a decreased expression during differentiation (Figure 3G).
3.6 Circ_0011269 affected osteogenic differentiation by sponging miR-122
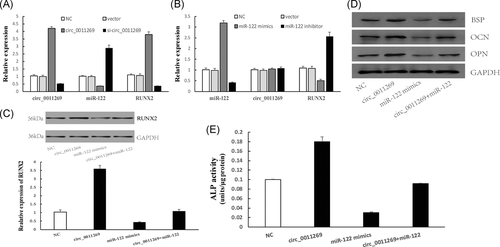
To further explore the role of circ_0011269 during osteogenic differentiation, hBMSCs were treated with conditions of circ_0011269 overexpression or si-circ_0011269, miR-122 overexpression or inhibitor and circ_0011269/miR-122 co-overexpression, respectively, transfection efficiency was measured in Figure S1A. The overexpression of circ_0011269 inhibited miR-122 expression and promoted RUNX2 expression, si-circ_0011269 showed the opposite results (Figure 4A). Overexpression of miR-122 could inhibit RUNX2 expression while had no effect on circ_0011269 expression (Figure 4B). In addition, RUNX2 expression in mRNA and protein level were barely changed after the co-overexpression of circ_0011269 and miR-122 (Figure 4C). Moreover, Western blot showed that the expression of osteogenic-associated biomarkers was notably upregulated by circ_0011269 overexpression and downregulated by miR-122 overexpression (Figure 4D). Besides, ALP activity was measured at 14th day of treated hBMSCs. As shown in Figure 4E, ALP activity was raised by circ_0011269 overexpression and barely changed by co-overexpression of circ_0011269 and miR-122.
4 DISCUSSION
In recent years, circRNAs were found to be widely expressed in human tissues and exhibited a tissue/disease-specific manner, which may serve as potential diagnostic and therapeutic targets for disease.5, 6 Thus, we focused on the expression profile and underlying mechanism of circRNAs in osteoporosis.
In this study, we measured the expression of circ_0011269 and circ_0011269-miRNAs regulatory network in osteoporosis for the first time. A total of 69 circRNAs were differentially expressed in osteoporosis patient samples compared to normal control samples according to microarray data. These circRNAs may provide valuable resources for osteoporosis research. Circ_0011269 was identified to be lowly expressed in osteoporosis patient samples by bioinformatics and qRT-PCR assay. Based on previous studies, circRNAs often act as sponges of miRNAs to regulate gene expression in physiological processes.19 For example, circMTO1 was found to suppress hepatocellular carcinoma progression via acting as the sponge of oncogenic miR-9.20 Circ_100395 regulated proliferation of lung cancer cells by modulating miR-1228/TCF21 pathway.21 The interactions of circ_0011269 and miRNAs were predicted by miRanda algorithm which based on sequence complementarity, binding energy and evolutionary conservation between miRNAs and the target sites. As a result, 38 miRNAs were predicted to be involved in the circ_0011269 interaction network. Among these enrolled miRNAs, miR-122 was mentioned to be abnormally expressed in osteoporosis in the previous study.22
Dual-luciferase reporter assay validated the target relationship of miR-122-circ_0011269 and miR-122-RUNX2, demonstrated that circ_0011269 may participate in osteoporosis by sponging miR-122. Thus, we focused on the potential axis of circ_0011269/miR-122/RUNX2 in regulating osteoporosis. qRT-PCR showed that during osteogenic differentiation of hBMSCs, the expression of circ_0011269 and RUNX2 were gradually increased while miR-122 expression decreased at 0, 7th, 14th and 21th day. Moreover, we noticed that osteogenesis differentiation was significantly elevated by overexpression of circ_0011269 but inhibited by miR-122 mimics. Besides, overexpression of circ_0011269 did not affect the expression of its linear form mRNA ZCCHC17 (Figure S1B).
In conclusion, the present study revealed circRNAs expression profile in osteoporosis and circ_0011269 may sponge 122 to promote osteoporosis progression, which may provide new insights for osteoporosis treatment.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
XX and FW designed the study and wrote the paper. XX, YC and BT performed data statistical analysis and qRT-PCR experiment. DW and ZY performed other experiments.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.