Retracted: Upregulation of acetylcholinesterase caused by downregulation of microRNA-132 is responsible for the development of dementia after ischemic stroke
Abstract
MicroRNA-132 (miR-132) has been shown to participate in many diseases. This study aimed to understand the correlation between the level of miR-132 and the severity of dementia post-ischemic stroke. An online tool (www.mirdb.org) was used to find the miR-132 binding site in acetylcholinesterase (ACHE) 3′-untranslated region (UTR), followed by a luciferase reporter assay to validate ACHE as a miR-132 target. A similar relationship between miR-132 and ACHE was also established in cerebrospinal fluid samples collected from human subjects. A negative correlation was established between ACHE and miR-132 by measuring the relative luciferase activity. Meanwhile, Western blot analysis and real-time polymerase chain reaction were also conducted to compare the levels of ACHE messenger RNA and protein between two groups (dementia positive, n = 26 and dementia negative, n = 26) or among cells treated with miR-132 mimics, ACHE small interfering RNA, and miR-132 inhibitors. As shown in the results, miR-132 can reduce the expression of ACHE. Further experiments were also carried out to study the effect of miR-132 and ACHE on cell viability and apoptosis, and the results demonstrated that miR-132 enhanced cell viability while suppressing apoptosis. In addition, ACHE reduced cell viability while promoting apoptosis. miR-132 targeted ACHE and suppressed its expression. Additionally, miR-132 and ACHE have been shown to affect the cell viability and apoptosis in the central nervous system.
1 INTRODUCTION
As demonstrated by many epidemiological studies, ischemic stroke is an important risk factor of dementia.1 Approximately one-fourth of elderly patients suffering from ischemic stroke were diagnosed with dementia within 3 months. The occurrence of dementia is associated with different clinical determinants, which include host features such as older age, a history of strokes and diabetes, and the location and size of present stroke.2 The elevated rate of adverse events in patients suffering from dementia stroke has led to the underestimation of the actual incidence of post-stroke dementia in prevalence studies.3
In severe dementia, the loss of central cholinergic neurons has been observed.4 However, no severe cholinergic neurodegeneration has been observed in early dementia, although the choline-like functions are impaired, possibly due to the change in receptor expression and the release of acetylcholine, which in turn leads to an unbalanced level of nerve growth factors.5 Acetylcholinesterase (ACHE) can inactivate cholinergic neurotransmission and its activity is reduced in the temporal cortex, hippocampus, and amygdala in the Alzheimer's dementia (AD) brain.6
Noncoding RNAs (ncRNAs) have been considered as promising molecules that can affect the prognosis of certain diseases. It was shown that the family of ncRNAs is indispensable in primary biological behaviors such as cell differentiation, inflammation, and apoptosis, although these ncRNAs were previously regarded as nonfunctional molecules when they were discovered more than 20 years ago.7 In particular, microRNAs (miRNAs) are the most extensively investigated members in the family of ncRNAs. These small molecules consist of 18 to 25 nucleotides and are often present in fragile chromosomal regions. They are able to trigger messenger RNA (mRNA) degradation and suppress translation.8 In fact, miRNAs are estimated to regulate as much as 60% of human genes.9 A number of processes are associated with miRNAs, including neuronal plasticity, synapse formation, cell differentiation, and neuronal development.10, 11 Therefore, it is not surprising to discover a close association between miRNAs and neurodegenerative diseases such as Parkinson disease,12 aggressive behaviors,13 depression,14 schizophrenia,15 and Alzheimer's disease.16
Dementia is associated with deregulated inflammatory signaling instead of an influx of inflammatory cells, as supported by the fact that many miRNAs related to immunity are consistently downregulated in dementia. In contrary, the upregulated activity of innate immunity is supported by the downregulation of miR-146b in cerebrospinal fluid (CSF). In addition, T lymphocytes are also involved in such pathways, even though a normal association between toll receptor signaling and monocytes was observed.17 Furthermore, miR-181a and miR-142-5p expression is reduced in CSF,18 while T cell sensitivity was demonstrated to be affected by miR-181.18 In addition, the expression of miR-191, miR-221, miR-142, and miR-15b was increased during T cell development but was downregulated in AD CSF.19
MicroRNA-132 (miR-132) has been reported to be downregulated in the central nervous system (CNS) of patients suffering from dementia,20 whereas ACHE was found to be upregulated in these patients.21 Using a miRNA online database, we identified AHCE as a target gene of miR-132. Hence, this study was carried out to clarify the correlation between miR-132 and ACHE in the development of dementia after ischemic stroke.
2 MATERIALS AND METHODS
2.1 Human sample collection
This study enrolled 52 subjects of ischemic stroke (26 with dementia and 26 without dementia) and all subjects diagnosed with ischemic stroke were placed into the “dementia positive” (n = 26, 18 males and 8 females aged 60-84 years old, with an average age of 71.5 years) group or the “dementia negative” (n = 26, 15 males and 11 females aged 64-81 years old, with a mean age of 70.3 years) group. Two milliliters of CSF was obtained from each patient. All CSF samples underwent 1200g centrifugation at 4°C for 5 minutes before use. The study protocol was in full compliance with international guidelines and was approved by our Institutional Ethics Review Board.
2.2 Quantitative polymerase chain reaction
Quantitative polymerase chain reaction (qPCR) was carried out to quantify RNA expression in sample tissues and cultured cells. In brief, the total RNA in CSF and serum samples was extracted using TRIzol (Invitrogen, Carlsbad, CA) and an RNAqueous-Micro Kit (Ambion, Austin, TX). The concentration of RNA extract was measured by a DR 6000 UV VIS ultraviolet spectrophotometer (HACH, Loveland, CO). Complementary DNAs were synthesized from RNA using a real-time PCR assay kit. qPCR was carried out with a SYBRGreen Master Mix (ABI, Foster City, CA) on a Realplex2 Master cycler (Eppendorf, Germany). The cycling condition was set according to kit instruction. The expression of ACHE mRNA and miR-132 was calculated using the method.
2.3 Cell culture and transfection
U87 cells and transfected cells were purchased from American Type Culture Collection (Rockville, MD) and incubated in a Roswell Park Memorial Institute-1640 medium containing 10% fetal bovine serum, streptomycin, and penicillin.
2.4 Vector construction and mutagenesis
The miR-132 binding site in ACHE 3′-untranslated region (UTR) was identified using TargetScan (targetscan.org). The sequence of ACHE 3′-UTR was then cloned into a pMIR-REPORTTM Luciferase Reporter Vector (Ambion, Austin, TX) to generate a luciferase construct containing the 3′UTR of wild-type ACHE. Meanwhile, the miR-132 binding site was mutated using a QuikChange Lightening Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA), and the mutant 3′UTR of ACHE was inserted to the same location in the pMIR-REPORT Luciferase Reporter Vector (Ambion, Austin, TX) to generate a luciferase construct containing the mutant 3′UTR of ACHE.
2.5 Cell proliferation assay
The viability of cells was measured using the Cell Counting Kit-8 (CCK-8) method (TransGen Biotech, Beijing, China). In brief, after transfection, the transfected cells and controls were seeded into 96-well (1000 cells per well) plates before conducting the CCK-8 assay. The frozen CCK-8 reagent was thawed and added to the plate (10 μg per well). After 2 hours of incubation, the count of live cells in each well was determined using the absorbance value at 450 nm.
2.6 Luciferase assay
U87 cells were plated into a 96-well plate and cultured overnight before the cells were cotransfected with miR-132 mimic/mimic control and wild-type/mutant reporter vector using a Lipofectamine 2000 system (Invitrogen). Luciferase activity of transfected cells was measured 1 day later using a Dual Luciferase kit (Promega, Madison, WI). Meanwhile, the luciferase activity of the renilla control was also detected as the endogenous control.
2.7 Western blot analysis
Western blotting was performed to determine whether miR-132 suppressed the expression of ACHE protein in U87 cells. In brief, the cultured cells and sample tissues were lysed and their total protein was extracted using a lysis buffer (Beyotime, Haimen, Jiangsu, China). The cell lysate was then centrifuged for 5 minutes at 14 000g and 4°C to remove cell debris using a QIAshredder spin column assembly (Qiagen, Hamburg, Germany). The protein samples were then resolved by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transblotted onto polyvinylidene difluoride membranes. The protein blots were then blocked with a blocking solution and incubated for 18 hours with the primary antibody (anti-ACHE antibody; 1:1000; SCBT, Santa Cruz, CA; anti-β-actin antibody; 1:10000; SCBT) at 4°C, followed by 1 hour of incubation at room temperature with a secondary antibody (1:1000; SCBT). Finally, the bound protein blots were visualized using an enhanced chemiluminescence system (PerkinElmer, Buckinghamshire, UK). An Alpha Imager was used to measure and calculate the intensity of each band on the blots.
2.8 Statistical analysis
Each experiment was performed for three times. The Student t test (two-tailed) was performed at a significant level of P < .05 (two-sided). Results were presented as mean ± SD. ANOVA was performed to analyze group differences. Differences in the demographic data between different groups were evaluated using the Student t test (continuous variables) or Pearson's χ2 test (categorical variables). P < .05 was deemed significant. All analyses were carried out using the SPSS 19.0 (IBM, Chicago, IL).
3 RESULTS
3.1 ACHE was a virtual target of miR-132
miR-132 has been implicated in many diseases. In this study, we aimed to understand the correlation between miR-132 and the severity of dementia after ischemic stroke. To achieve this goal, an online tool was used to locate the regulatory gene of miR-132, and ACHE was then found as a miR-132 target (Figure 1). Furthermore, to clarify the correlation between miR-132 and ACHE, a luciferase reporter assay was done by cotransfecting cells with miR-132 and wild-type/mutant ACHE 3′UTR. The luciferase activity of wild-type ACHE 3′UTR was decreased significantly (Figure 2) by miR-132 (P < .05 vs scramble control), whereas the cells cotransfected with miR-132 and mutant ACHE 3′UTR showed a luciferase activity comparable to that in cells transfected with the control miRNA (Figure 2). These results confirmed the role of ACHE as a miR-132 target. Furthermore, the relationship between miR-132 and ACHE mRNA expression was analyzed using tissue samples (n = 52), and the results demonstrated a negative correlation (Figure 3).

ACHE as the candidate target gene of miR-132 in cells with the “seed sequence” in the 3′UTR. ACHE, acetylcholinesterase; UTR, untranslated region
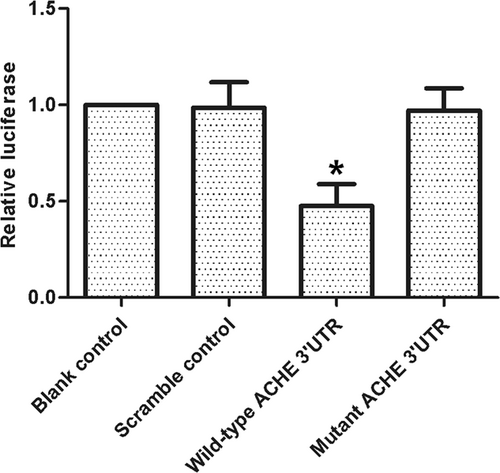
Luciferase activity reporter assay was conducted to verify ACHE as the direct target gene of miR-132, luciferase activity of wild-type but not mutant ACHE 3′UTR in cell transfected with miR-132 was much higher (*P < .05 vs scramble control). ACHE, acetylcholinesterase; UTR, untranslated region
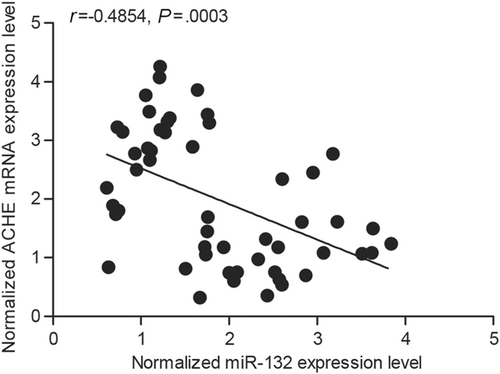
The correlation between the expression level of miR-132 and ACHE mRNA between dementia positive and dementia negative, which was confirmed the correlation of miR-132 and ACHE was negative with negative correlation coefficient being −0.3697 (dementia positive, n = 26 and dementia negative, n = 26). ACHE, acetylcholinesterase; mRNA, messenger RNA
3.2 Expression of miR-132 and ACHE in tissues from different groups
Tissues collected from two groups (dementia positive, n = 26 and dementia negative, n = 26) were studied to further explore the correlation between miR-132 and ACHE 3′UTR. The results demonstrated that the expression of miR-132 decreased in the dementia positive group (Figure 4A) as compared with that in the dementia negative group (P < .05 vs dementia negative), whereas the expression of ACHE mRNA (Figure 4B) increased in the dementia positive group as compared with that in the dementia negative group (P < .05 vs dementia negative). Through densitometry analysis, it was found that the expression of ACHE protein (Figure 4C) was increased in the dementia positive group as compared with that in the dementia negative group. Furthermore, the expression of ACHE mRNA/protein in CSF was measured by transfecting the cells with scramble control, miR-132 mimics, ACHE siRNA, or miR-132 inhibitors. Figure 5 showed that the expression of ACHE mRNA (bottom panel) in the cells transfected with miR-132 mimics and ACHE siRNA was significantly lower than that in the cells transfected with the scramble control (P < .05 vs scramble control), whereas the expression of ACHE mRNA in the cells transfected with miR-132 inhibitors was apparently higher than that in the cells transfected with the scramble control (P < .05 vs scramble control), confirming the presence of a negative regulatory correlation between miR-132 and ACHE.
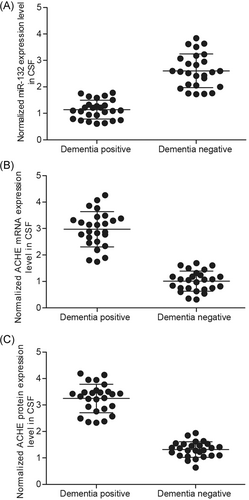
The expression of miR-132 decreased in dementia positive (A) compared with dementia negative group (P < .05 vs dementia negative group), while the expressions of ACHE mRNA (B) and protein (C) increased in dementia positive group compared with dementia negative group (P < .05 vs dementia negative group). ACHE, acetylcholinesterase; CSF, cerebrospinal fluid; mRNA, messenger RNA
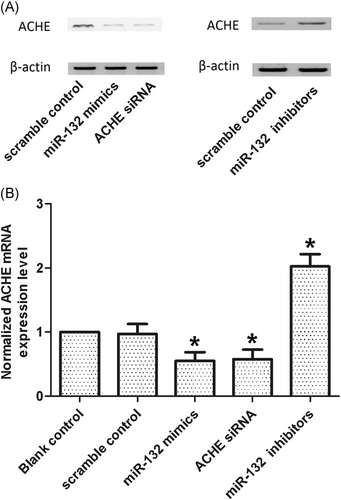
When transfected with the cells with scramble control, miR-132 mimics, ACHE siRNA, and miR-132 inhibitors, the expressions level of ACHE protein (A) and mRNA (B) treated with miR-132 mimics and ACHE siRNA decreased (*P < .05 vs scramble control), while ACHE mRNA and protein levels of cells treated miR-132 inhibitors increased (*P < .05 vs scramble control). ACHE, acetylcholinesterase; mRNA, messenger RNA; siRNA, small interfering RNA
3.3 miR-132 and ACHE affected cell viability
The relative cell viability was also assayed by transfecting the cells with scramble control, miR-132 mimics, ACHE siRNA, or miR-132 inhibitors. The cells transfected with miR-132 inhibitors demonstrated evidently reduced viability (Figure 6A) as compared to the cells transfected with the scramble control (P < .05 vs scramble control), whereas the cells treated with miR-132 mimics and ACHE siRNA demonstrated significantly higher viability (P < .05 vs scramble control), indicating that miR-132 enhanced the viability of cells, whereas ACHE reduced cell viability.
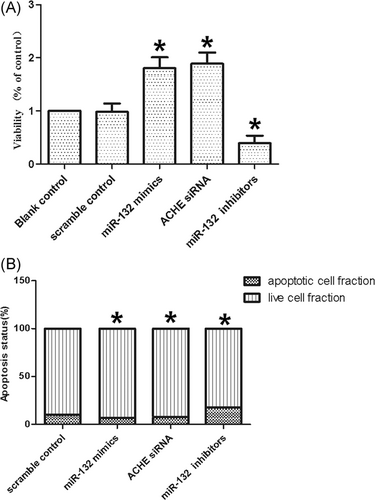
A, Cells transfected with miR-132 inhibitors showed evidently downregulated viability (*P < .05 vs scramble control), while cells transfected with miR-132 mimics and ACHE siRNA showed comparably higher viability (*P < .05 vs scramble control). B, Cells transfected with miR-132 mimics and ACHE siRNA inhibited cell apoptosis (*P < .05 vs scramble control), while cells transfected with miR-132 inhibitors accelerated cell apoptosis (*P < .05 vs scramble control). ACHE, acetylcholinesterase; siRNA, small interfering RNA
3.4 miR-132 and ACHE affected cell apoptosis
Subsequently, cell apoptosis was measured after the cells were treated with the scramble control, miR-132 mimics, ACHE siRNA or miR-132 inhibitors. After transfection with miR-132 mimics and ACHE siRNA, the number of apoptotic cells became lower (P < .05 vs scramble control), whereas the transfection with miR-132 inhibitors increased the number of apoptotic cells (P < .05 vs scramble control), indicating that miR-132 suppressed apoptosis, whereas ACHE promoted apoptosis.
4 DISCUSSION
As a class of small RNAs, miRNAs can bind to protein-coding mRNAs to regulate gene expression. miR-132, a miRNA enriched in neurons, participates in various physiological processes including angiogenesis, inflammation, synaptic transmission, and neurological development.22 In studies of Miller et al23 and Wanet et al,22 a series of neurological diseases, such as tauopathies, Parkinson's disease, and Alzheimer's disease, were attributed to the dysregulation of miR-132, indicating that miRNA has a more extensive influence on various brain diseases. Diverse neuronal activities, including neuron differentiation, apoptosis, and survival, are closely linked to brain-derived neurotrophic factors. Remenyi et al showed that brain-derived neurotrophic factor exerted a favorable effect on CNS neurons by increasing the level of miR-132 expression.24 In this study, an online database of miRNA was searched to confirm ACHE as a target gene of miR-132. Subsequently, using luciferase assays, ACHE was validated to be a direct target of miR-132. Additionally, a negative regulatory relationship was established between miR-132 and ACHE in CSF collected from patients. Moreover, we found that the expression of ACHE mRNA was significantly reduced in cells transfected with miR-132 mimics and ACHE siRNA, whereas the transfection with miR-132 inhibitors significantly increased the mRNA expression of ACHE, confirming the negative correlation between ACHE and miR-132.
Radić et al25 found that ACHE contributed to the discontinuation of cholinergic transmission, also known as the enzymatic breakdown of ACh. As one of the fastest enzymes in the body, ACHE degrades ACh at a speed of approximately 25 000 ACh molecules per second.26 Located on chromosome 7 with a length of seven kilobases, the gene of ACHE contains several exons, ie, E2, E3, and E4, encoding a core structure of ACHE enzyme. Taylor and Radic27 showed that the pre-mRNA of ACHE was predisposed to alternative splicing, which resulted in the generation of three posttranscriptional species. Several studies28 showed that the posttranslational processing and function of ACHE were determined by the C-terminal domains of the diverse splice variants.29, 30 The major neuropathological alterations correlated with dementia include substantial synaptic and neuronal loss, neurofibrillary tangle formation, and b-amyloid plaque buildup in crucial brain areas. In particular, evidence has pointed to cholinergic dysfunction in dementia brains.
It was reported that ACHE expression was reduced in the brain of Alzheimer's disease.31 In addition, ACHE expression was elevated in β-amyloid plaques32 by participating in the fibrillogenesis of β-amyloid.33 In particular, the investigators were interested in the expression of different ACHE species in the brain of patients with AD, who showed a reduced level of G4 tetramer and a slightly increased level of G1 monomer and G2 dimer.34 Intriguingly, most severely affected cases of AD showed enhanced activity of these light forms.35 Here, we compared CSF samples of two groups (dementia positive, n = 26 and dementia negative, n = 26) and demonstrated that the dementia positive group had a reduced level of miR-132 expression (Figure 4A) along with an increased level of ACHE expression (Figure 4B). Furthermore, the relative viability of cells was also measured by transfecting the miR-132 mimics, ACHE siRNA or miR-132 inhibitors. The results demonstrated that miR-132 inhibitors significantly reduced cell viability (Figure 6A), whereas miR-132 mimics and ACHE siRNA significantly increased cell viability. The cell type we used in this study was U87MG cell line. It was reported that the current U87MG cell line has undergone certain changes in the past.36 We realized this point after being pointed out during review process, and since we just chose a glioma cell line to confirm the regulatory association between miR-132 and ACHE, the unknown origin does not influence the conclusion of this study.
In conclusion, the findings of the present study showed that ACHE expression was targeted and suppressed by miR-132. In addition, downregulation of microRNA-132 increased the expression of ACHE, which was responsible for the pathogenesis of dementia upon ischemic stroke, at least partially by regulating cell apoptosis in the CNS.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
F-wY planned and directed the study; F-wY, HW, CW, and G-nC collected, analyzed, and visualized the data; F-wY completed the manuscript and GC improved the final manuscript. All authors read and approved the final manuscript.