Research on the roles of genes coding ATP-binding cassette transporters in Porphyromonas gingivalis pathogenicity
Abstract
Porphyromonas gingivalis, as a major pathogen of periodontitis, could rapidly adhere to and invade host gingival epithelial cells (GECs) for the induction of infection. One ATP-binding cassette (ABC) transporter gene was found to be upregulated during this infection process, however, the molecular mechanisms remain unclear. In this study, we systemically investigated the messenger RNA level changes of all ABC transporter family genes in P. gingivalis while being internalized within GECs by real-time polymerase chain reaction. We identified that two ABC transporter genes, PG_RS04465 (PG1010) and PG_RS07320 (PG1665), were significantly increased in P. gingivalis after coculturing with GECs. Mutant strains with knockout (KO) of these two genes were generated by homogenous recombination. PG_RS04465 and PG_RS07320 KO mutants showed no change in the growth of bacteria per se. Knockdown of PG_RS07320, but not PG_RS04465, caused decreased endotoxin level in the bacteria. In contrast, both mutant strains showed decreased Arg- and Lys-gingipains activities, with significantly reduced adhesion and invasion capabilities. Secreted interleukin-1β (IL-1β) and IL-6 levels in GECs cocultured with PG_RS04465 or PG_RS07320 KO mutants were also decreased, whereas, only the cells cocultured with PG_RS07320 KO mutants showed significant decrease. In addition, virulence study using mouse revealed that both KO mutant strains infection caused less mouse death than wild-type strains, showing reduced virulence of two KO strains. These results indicated that ABC transporter genes PG_RS04465 and PG_RS07320 are positive regulators of the virulence of P. gingivalis.
1 INTRODUCTION
Porphyromonas gingivalis, a Gram-negative anaerobe, has been established as one major pathogen of chronic periodontitis.1 P. gingivalis can rapidly attach to and invade gingival epithelial cells (GECs), which are the first line of afflicted host cells within 30 minutes during P. gingivalis infection.2 The major fimbriae of P. gingivalis are not only involved in adhesion to GECs,3 but also trigger the subsequent signal transduction events associated with invasion. The binding of fimbriae to the β1 integrin receptor leads to the activation of paxillin and focal adhesion kinase, consequently resulting in the remodeling of the actin cytoskeleton.4, 5
Upon infection by P. gingivalis, multiple signaling cascades within GECs controlling the transcription of cytokines associated with immune response and inflammatory reactions have been shown to be activated, including the interleukin-1β (IL-1β), IL-6, IL-33, and tumor necrosis factor-α.6-8 Interestingly, P. gingivalis is capable of replication at a high speed inside GECs and remain viable for an extended period.9, 10 Arg-gingipains (RGP) and Lys-gingipains (KGP) are two major cysteine proteinases produced by P. gingivalis, which serve as two vital virulence factors.11, 12 In addition, previous proteomics and genomics studies revealed that a number of functional genes and proteins are closely associated with P. gingivalis infection by regulating the adaptation and survival of P. gingivalis in GECs,13-16 such as ATP-binding cassette (ABC) transporters.17
ABC transporters, as one of the largest transmembrane protein superfamilies, are highly conservative among all species. As ATP-dependent pumps, they transport a diversity of substances across biological membranes using ATP as energy source.18-20 Eukaryotic ABC transporters play important roles in the import of essential nutrients, the export of signaling molecules and toxins, and multidrug resistance in cancer cells.21 Previous investigations also showed that ABC transporters were required for bacterial viability, virulence, and pathogenicity, depending on the substrates they transport.22, 23 ABC transporters were also shown to be involved in multiple colonization processes, including heme uptake and use, formation and morphology of biomembranes, bacterial adhesion and colonization, and interaction between bacteria.24 For instance, bacteria exploited mammalian extracellular glycosaminoglycans (GSGs) as a target to establish colonization, and ABC transporters mediated GSG import in the pathogenic Streptobacillus moniliformis.25 Moreover, ABC transporters participated in the bacterial uptake of different nutrients, including Fe2+, amino acids, vitamins, and oligopeptides, thus contributing to bacteria survival in the host microenvironment.
By whole-genome analysis, 40 ABC transporter genes are identified in P. gingivalis strain W83.26 The functions of these ABC transporter genes in the virulence of P. gingivalis have not been systemically investigated yet. A previous study showed that the tlr gene, previously described as an indispensable gene for P. gingivalis growth at low hemin concentration, is immediately downstream of four open reading frames (htrABCD).27 The htrABCD frames encode a putative ABC transporter system with a sequence similar to heme transport systems of other bacteria, suggesting that ABC transporters play roles in P. gingivalis virulence. Another report using DNA microarray to compare gene expression of P. gingivalis biofilm cells and counterpart planktonic cells showed that six ABC transporter genes were upregulated and might regulate biofilm growth.28 Consistently, during P. gingivalis infection in GECs, one ABC transporter gene was increased and involved in bacterial invasion.17 The altered expression of ABC transporter genes such as PG0258 and PG1010 in P. gingivalis associated with periodontitis was also indicated by other genome microarray assays.29, 30 However, the exact molecular mechanisms remain elusive.
In this study, we aimed to explore the specific mechanisms by which ABC transporters regulate the P. gingivalis infection process. Real-time polymerase chain reaction (PCR) was used to detect P. gingivalis ABC transporter genes differentially expressed after prolonged incubation with GECs. The roles of PG_RS04465 (PG1010) and PG_RS07320 (PG1665) in the virulence of P. gingivalis were further investigated by the establishment of mutant strains with the knockout of these two genes.
2 MATERIALS AND METHODS
2.1 Bacterial strains and growth conditions
P. gingivalis W83 and its derivatives were anaerobically maintained in blood agar plates and grown in brain heart infusion (BHI) liquid medium (Oxiod, Newport, England), supplemented with 5 mg/mL hemin and 5 mg/mL vitamin K under anaerobic atmosphere.
2.2 Isolation and culture of gingival epithelial cells
Healthy human gingival tissue samples were obtained with informed consent from periodontal healthy adults undergoing third molar extraction at the Oral Surgery Clinic of Hospital of Stomatology, Sun Yat-Sen University. GEC cells were prepared for culture as previously described.31 The authenticity of GECs was confirmed by cell morphology and immunohistochemistry with monoclonal antibodies against human pankeratin. The isolated GECs were cultured in serum-free keratinocyte medium (K-SFM), supplemented with keratinocyte growth factors, hydrocortisone, bovine insulin, bovine pituitary extract, gentamicin sulfate, amphotericin B, and 0.15 mM CaCl2. GEC cells undergoing 5-10 passage were used for functional studies.
2.3 RNA extraction and real-time polymerase chain reaction
P. gingivalis was cultured under normal conditions or cocultured with GECs for 18 hours. After washing with PBS, cells were lysed with lysis buffer containing 0.5 mg/mL DNase I, 0.25 mg/mL RNase A, and 1 mM MgCl2 to remove the eukaryotic DNA and RNA. Total RNA was extracted from P. gingivalis using the RiboPure-Bacteria Kit (Ambion, Austin, Texas) according to the manufacturer's instructions and stored at −80°C. The complementary DNA (cDNA) was synthesized by reverse transcription using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA) following the manufacturer's instructions. Real-time PCR was conducted on a Bio-Rad CFX96 Real-time PCR system using the Takara SYBR Premix Ex Taq II.
2.4 Generation of Porphyromonas gingivalis gene knockout mutant strains
P. gingivalis mutants with knockout of PG_RS04465 or PG_RS07320 genes were constructed using homologous recombination using the pShuttle-ErmF-ErmAM vector (Takara, Shiga, Japan). In brief, genomic fragments of each gene containing tetracycline were amplified by PCR and ligated with pMD-18T vector, which was then used for the construction of pShuttle-HA up-HA down-ErmF-ErmAM vector. The recombinant HA up-HA down-errnF-ermAM vector for the induction of homologous recombination was obtained by the digestion of pShuttle-HA up-HA down-ErmF-ErmAM vestor using Xhol enzyme. The recombinant plasmids were then introduced into P. gingivalis by electroporation and selected on tryptic soy broth (TSB) agar plates supplemented with tetracycline. Specifically, 200 ng recombinant vectors were introduced into 200 uL P. gingivalis resuspended in electroporation buffer by electroporation, followed by further culture in 1 mL fresh liquid culture medium for 24 hours, and screened by culture on agar plates with 5 mg/L clindamycin for 7 to 10 days. Positive colonies were verified by PCR and sequencing (Life Technologies, Gaithersburg, MD). Primers used for mutant establishment or screening are listed as follows (Table 1).
| Genes | Primer | Primer Sequence | Product, bp |
|---|---|---|---|
| PG_RS04465 | F-up | CCCGGGGATCCTCTAGAGAT CGAAGTGCTGATAGAGGGCTTCAG | 1045 |
| R-up | CACTTCGCCCATTGTAGAGCCTCGAG GTCCGAGCAGTCCCGTAATACTC | ||
| F-down | GGCAAGATAGACGAGACGGACTCGG GCTCTACAATGGGCGAAGTGGGTG | 750 | |
| R-down | CTATCGGGGGTACCGCTAGC AGTGGTGCCTCGTACATTGATGCAG | ||
| PG_RS07320 | F-up | CCCGGGGATCCTCTAGAGAT CGGCTGATGTCTTATTCGGGGAAAG | 975 |
| R-up | ACCGCCGGCTATGATGATAACTCGAG GCCCGATACCTCACGTAGAATACC | ||
| F-down | TTCTACGTGAGGTATCGGGCCTCGAG TTATCATCATAGCCGGCGGTATAC | 847 | |
| R-down | CTATCGGGGGTACCGCTAGC TCCGTCTCGTCTATCTTGCCGACA | ||
| ErmF-ErmAM | F | TCAATGTACGAGGCACCACT GCTAGCGGTACCCCCGATAGCTTCC | 2197 |
| PG_RS04465 | R | TGCATGCCTGCAGGTCGACG GAATTCGAGCTCCTGCAGGTCGACT | |
| ErmF-ErmAM | F | GGCAAGATAGACGAGACGGA GCTAGCGGTACCCCCGATAGCTTC | 2197 |
| PG_RS07320 | R | TGCATGCCTGCAGGTCGACG GAATTCGAGCTCCTGCAGGTCGA |
- Abbreviations: F, forward; R, reverse
2.5 Growth curve of P. gingivalis
P. gingivalis W83 and KO mutant strains were anaerobically grown in TSB liquid medium at 37°C. Bacterial growth curve was drawn by determination of optical density (OD) 600 at 2, 4, 8, 12, 16, 22, 28, 32, and 48 hours, respectively, using an ultraviolet-visible spectrophotometer.
2.6 Endotoxin level measurement
The endotoxin (lipopolysaccharide; LPS) level was detected using the limulus amebocyte lysate assay. P. gingivalis W83 and KO mutant strains were collected and the LPS level was determined using Limulus PS Single Test Kit (Wako Pyrostar, Saitama, Japan) following the manufacturer's instructions.
2.7 Invasion assay
GECs were seeded into 24-well plates until the confluence reached 50%. P. gingivalis W83 or KO mutant strains were added into cells and incubated for 36 hours. Subsequently, GECs were washed, lysed, and centrifuged at 1000g for 5 minutes. The invaded bacteria within the supernatants were collected and diluted to seed on the TSB agar plates. The number of colonies from each stain was counted for evaluation of the number of invaded bacteria.
2.8 Adherence assay
GECs were first fixed with 5% buffered formalin to inhibit P. gingivalis invasion, and incubated with P. gingivalis W83 or KO mutant strains for 30 minutes at 37°C. After being washed five times with PBS to remove nonadherent bacteria, GECs were lysed and centrifuged at 1000g for 5 minutes. The suspended bacteria were diluted and seeded onto TSB agar plates for colony counting as described in the invasion assay.
2.9 Enzyme activity
Arg-gingipains (RGP) and Lys-gingipains (KGP) protease activities were determined by the enzymatic activity assay using substrates N-α-benzoyl-dl-arginine-p-nitroanilide (BRpNA) and N-α-acetyllysine-p-nitroanilide (AcKpNA), respectively, as described previously.32 The increases of absorbance at 405 nm due to p-nitroaniline were monitored for calculation of relative enzyme activity through division by the total protein concentration measured by Bradford protein assay.
2.10 Cytokine level detection
GECs and P. gingivalis were cocultured for 6 and 24 hours, respectively, which were then subjected to centrifugation for the removal of bacteria. Culture supernatants were analyzed for IL-6 and IL-1β levels using enzyme-linked immunosorbent assay (ELISA) Kit (R&D, Minneapolis, MN). Data were normalized by subtraction of OD 450 in blank wells.
2.11 Virulence studies
In total, 80 BALB/c mice were randomly divided into four groups (n = 20) and subcutaneously injected with saline, P. gingivalis W83, or mutant strains (at a dose of 2 × 1010 c.f.u per mouse), respectively. Mice health status and death were then examined on days 2, 4, and 8, for drawing the survival curve.
2.12 Statistical analysis
Statistical analysis was performed using GraphPad Prism 5 software (GraphPad Software Inc, San Diego, CA). Results were presented as mean ± SD or mean ± SEM and statistical significance was assessed by the Student two-tailed t test. Significant differences are expressed as follows: *P < 0.05, **P < 0.01, ***P < 0.001.
3 RESULTS
3.1 Identification of differentially expressed ATP-binding cassette transporter genes
To investigate the roles of P. gingivalis ABC transporter genes in the infection process, P. gingivalis was cultured alone or cocultured with GECs for 18 hours to identify the differentially expressed genes by real-time PCR. Among 40 ABC transporter genes, distinct differences were observed between control and intracellular bacteria after internalizing within GECs (Figure 1A and 1B). The messenger RNA (mRNA) levels of three ABC transporter genes were significantly upregulated, while those of 15 ABC transporter genes were significantly decreased. Interestingly, the expression of six genes exhibited more than two-fold changes during infection. ABC transporter genes PG_RS04465 (previously known as PG1010) and PG_RS07320 (previously known as PG1665) showed dramatic increases (3.4- and 2.4-folds, respectively), indicating their essential roles in P. gingivalis invasion process. Besides, these two genes were supposed to be ABC transporter ATP-binding proteins, which were therefore selected for further studies. Genomic sequences of PG_RS04465 and PG_RS07320 genes, as well as their protein sequences, are shown in Table S1. These two proteins shared high homology with other ABC transporter proteins in P. gingivalis and other species (Tables S2 and S3).
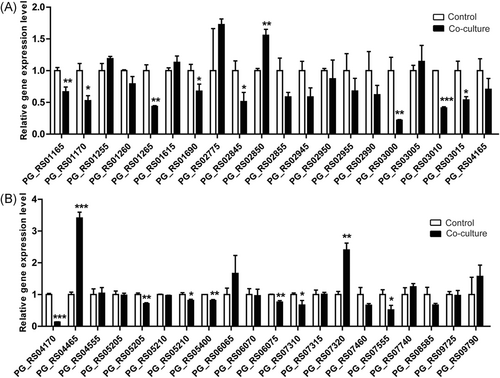
The messenger RNA (mRNA) level changes of Porphyromonas gingivalis ATP-binding cassette (ABC) transporter genes during invasion into gingival epithelial cells (GECs). A,B, P. gingivalis was cultured alone or cocultured with GECs for 18 hours. The mRNA levels of P. gingivalis ABC transporter family genes were assessed by real-time polymerase chain reaction (PCR). Figure 1A and 1B was a continuation of the same dataset obtained by RT-PCR. *P < 0.05, **P < 0.01, ***P < 0.001
3.2 Construction of P. gingivalis mutant strains
To explore the functions of ABC transporter genes, PG_RS04465 and PG_RS07320, in the infection process of P. gingivalis, we generated mutant strains with knockout (KO) of PG_RS04465 or PG_RS07320 gene by insertional mutagenesis. P. gingivalis KO mutant strains were verified by sequencing and designated as KO_PG_RS04465 and KO_PG_RS07320, hereafter.
3.3 Reduced virulence factors in P. gingivalis KO mutant strains
Since ABC transporter genes PG_RS04465 and PG_RS07320 were significantly increased after incubation with GECs for 18 hours, we suppose they might regulate the virulence factors in P. gingivalis. By monitoring growth, we revealed that the growth of P. gingivalis was not significantly affected by the knockout of PG_RS04465 or PG_RS07320 gene (Figure 2A). Moreover, KO_PG_RS04465 did not show significant alteration of endotoxin level, while the knockout of PG_RS07320 gene led to significantly reduced endotoxin level in P. gingivalis (Figure 2B).
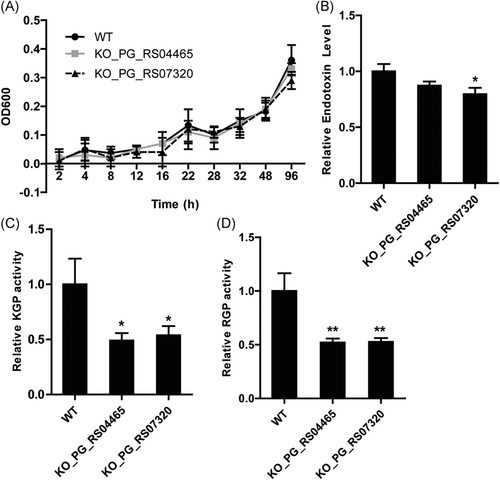
P. gingivalis knockout (KO) mutant strains showed reduced virulence factors. A, Growth curves of the wild-type (WT) and KO mutant P. gingivalis strains. WT P. gingivalis, mutant strains KO_PG_RS04465 and KO_PG_RS07320 were grown anaerobically and their growth was determined by measuring optical density 600 at indicated time points. B, Relative endotoxin levels in WT and KO mutant P. gingivalis strains. C, Relative Arg-gingipain (RGP) activities in WT and KO mutant P. gingivalis strains. D, Relative Lys-gingipain (KGP) activities in WT and KO mutant P. gingivalis strains. *P < 0.05, **P < 0.01
Arg- (RGP) and Lys-gingipains (KGP) are two cysteine proteinases produced by P. gingivalis, which act as major virulence factors in the infection process.11, 12 We also detected the enzyme activities of RGP and KGP in wild-type W83 strain and two KO mutant strains. As shown in Figure 2C and 2D, the enzymatic activities of both RGP and KGP were decreased by about 50% in mutant strains KO_PG_RS04465 and KO_PG_RS07320, compared with the wild-type strain, suggesting the reduced virulence ability in mutant strains. Collectively, these results indicated that the knockout of ABC transporter genes, PG_RS04465 and PG_RS07320, did not affect the growth of bacteria per se, but exerted inhibitory effects on virulence factors including RGP and KGP.
3.4 P. gingivalis KO mutant strains exhibited decreased adhesion and invasion properties
Given the involvement in the regulation of virulence factors, the effects of PG_RS04465 and PG_RS07320 KO mutations on bacterial adhesion and invasion were further analyzed. In the adhesion assay, we found that the adhesion capabilities of mutant strains KO_PG_RS04465 and KO_PG_RS07320 to GECs were significantly suppressed compared with the wild-type W83 (Figure 3A). Moreover, the number of mutant strain cells invading inside GECs was quantified and compared with the parental strains. We observed that the invasive ability of KO_PG_RS04465 and KO_PG_RS07320 strains was also significantly decreased in comparison to the wild-type strains. It is widely known that GECs exert immune response to P. gingivalis invasion by secreting numerous cytokines such as IL-1β and IL-6. Thus, we next analyzed IL-1β and IL-6 levels in the supernatants of GECs by ELISA method. A slight decrease of IL-1β and IL-6 secretion was observed in GECs in response to KO_PG_RS04465, compared with wild-type W83, however, the difference was not significant (P > 0.05; Figure 3C and 3D). GECs infected by KO_PG_RS07320 secreted significantly decreased IL-1β and IL-6, indicating that KO_PG_RS07320 strain possessed reduced virulence compared with the wild-type strain. In summary, these results demonstrated that these two ABC transporter gene KO mutant strains showed decreased adhesion and invasion capabilities against GEC cells.
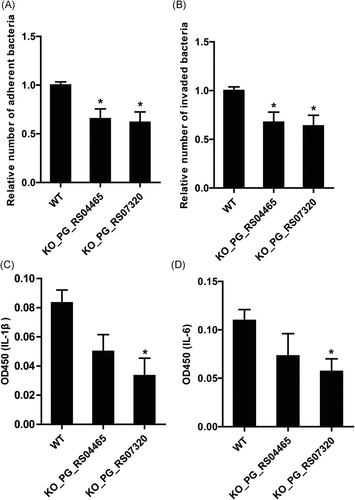
Adhesion and invasion properties of P. gingivalis KO mutant strains. A, Relative numbers of adherent bacteria onto GECs. B, Relative numbers of invaded bacteria within GECs. C, Interleukin-1β (IL-1β) levels of GECs in response to WT and KO mutant P. gingivalis strains. D, IL-6 levels secreted by GECs in response to WT and KO mutant P. gingivalis strains. *P < 0.05. GEC, gingival epithelial cell; KO, knockout; WT, wild-type
3.5 P. gingivalis KO mutant strains showed decreased virulence
To further evaluate the virulence differences between P. gingivalis wild-type and KO mutant strains, an animal study was further carried out. Mice were subcutaneously injected with saline, P. gingivalis W83 wild-type, KO_PG_RS04465, and KO_PG_RS07320 at a dose of 2 × 1010 c.f.u per mouse, followed by close monitoring of mouse health status and death. All mice in the saline group remained healthy till the end of the virulence experiment as expected, while 12 of total 20 mice died 8 days after infection of P. gingivalis W83 wild-type strain (Figure 4). Meanwhile, mice infected with mutant strains, KO_PG_RS04465 and KO_PG_RS07320, showed a significantly reduced number of mouse death, compared with those infected with the wild-type strains (Figure 4), suggesting less virulence ability of the two KO mutant strains in contrast to the wild-type strain.
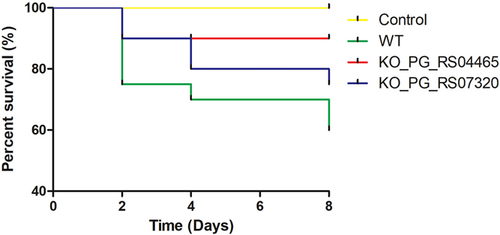
Reduced virulence of P. gingivalis KO mutant strains. BALB/c mice were subcutaneously injected with saline, P. gingivalis W83, and mutant strains with knockout of PG_RS04465 and PG_RS07320 genes (n = 20), respectively, and percentages of mouse survival were monitored at days 2, 4, and 8 after infection for drawing the survival curve. KO, knockout
4 DISCUSSION
P. gingivalis could rapidly attach to and invade the GECs, and fimbriae and cysteine proteinases such as RGP and KGP are well-known virulence factors, contributing to the P. gingivalis infection process.33 It has also been revealed that one ABC transporter gene was significantly increased in P. gingivalis during infection process, however, the underlying molecular mechanisms are poorly understood. In this study, we performed a real-time PCR assay to screen differentially expressed P. gingivalis ABC transporter genes during the invasion process. In total, 18 ABC transporter genes were significantly regulated, and expression of six ABC transporter genes exhibited more than two-fold changes.
PG_RS04465 and PG_RS07320, as the two most significantly upregulated ABC transporter genes, were selected for further studies. Mutant strains with knockout of PG_RS04465 or PG_RS07320 gene were constructed by homologous recombination technique. Knockout of PG_RS04465 and PG_RS07320 genes caused greatly reduced enzyme activities of Arg- and Lys-gingipains, two cysteine proteinases produced by P. gingivalis as major virulence factors, however, the bacteria growth was not influenced by the knockout of these two genes. Consistently, KO strains exhibited reduced capabilities in adhesion and invasion. Furthermore, our animal experiment revealed that infection with KO mutant strains caused less mouse death than the wild-type strains, suggesting reduced virulence ability of P. gingivalis with knockout of PG_RS04465 and PG_RS07320 genes. Collectively, these results indicated the involvement of these two ABC transporter genes, PG_RS04465 and PG_RS07320, in the virulence of P. gingivalis.
ABC transporters were associated with the transport of various substances, including import of nutrients and ions, secretion of toxins and signaling molecules, and even multiple anticancer drugs. These ABC transporters play fundamental and important roles in many physiological and pathological processes, mainly depending on their capabilities of transporting other bioactive molecules. For instance, the chvE-gguAB gene, an ABC transporter gene associated with glucose and galactose uptake, was identified as a regulator of the virulence of Agrobacterium tumefaciens.34 Prokaryotic ABC transporters were also shown to regulate cell-to-surface and/or cell-to-cell interactions, antibiotic resistance, and biofilm morphology.35 Moreover, ABC transporters are responsible for the transport of Fe2+, thus regulating the bacterial survival in the host microenvironment. Thus, we speculated that the regulation of ABC transporter expression might influence the virulence of P. gingivalis, since P. gingivalis has an obligate requirement for iron, preferably in the form of heme or its oxidized product, hemin. In the present study, we demonstrated that the ABC transporter genes, PG_RS04465 and PG_RS07320, are needed for the adhesion and invasion of P. gingivalis to GECs, which are initial steps during infection. These results supported the hypothesis that bacterial ABC transporters are essential for cell virulence and pathogenicity of P. gingivalis. Furthermore, the possible involvement of the transport of bioactive molecules such as heme in this process deserves further investigations.
In functional studies, strains KO_PG_RS04465 and KO_PG_RS07320 were shown to have similar properties in most assays done in this study. However, slight differences remain between two groups. For example, KO_PG_RS07320 resulted in weaker immune response than wild-type, exemplified by less section of IL-1β and IL-6, but this is not the case for KO-PG_RS04465. These results suggested that the functions of these two ABC transporter genes are not exactly the same. In the real-time PCR screening, we also identified several genes significantly downregulated, one of which was chosen as a target gene for mutagenesis. This gene was initially proposed as a negative regulator of adhesion and invasion in P. gingivalis into GECs. Interestingly, preliminary data showed that knockout of this gene does not affect the adhesion and invasion capabilities (data not shown). We supposed its effect was compromised by other positive regulators of this process, perhaps caused by other ABC transporter genes. These results also suggested that the infection process is exquisitely regulated by ABC transporter family genes. Therefore, the roles of other differentially expressed ABC transporter genes in the infection processes merit further exploration.
Our analysis showed that both Arg-gingipains (RGP) and Lys-gingipains (KGP) levels in P. gingivalis were greatly decreased by the knockout of PG_RS04465 and PG_RS07320 gene. The protease activity was previously shown to regulate activation of human oral epithelial cell surface receptors and induction of IL-6 secretion, which was closely associated with P. gingivalis virulence and infection.36, 37 Thus, it is reasonable to speculate that changes of the protease activity might serve as a molecular mechanism underlying the influence of virulence by PG_RS04465 and PG_RS07320 gene expression. In addition, a previous investigation showed that the nuclear factor-κB (NF-kB) and mitogen-activated protein kinase pathways in oral epithelial cells could mediate P. gingivalis infection.38 The possible involvement of NF-kB signaling and other pathways in the regulation of P. gingivalis pathogenicity deserve further investigations. In conclusion, the results indicate that PG_RS04465 and PG_RS07320, the ABC transporter genes, are required for the virulence abilities of P. gingivalis.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (Funding No. 81300887) and Natural Science Foundation of Guangdong Province, China (Funding No. 2018A030313674).
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
LG designed and performed the main part of the experiments; CZ designed the study and wrote the paper; YM performed the mutant construction experiment; XL and LZ took part in the in vitro and in vivo virulence experiments; CZ and QC collected and analyzed the data. All authors read and approved the final manuscript.