Enhancing the dung beetle iDNA tool for mammalian biodiversity monitoring and ecological studies
优化粪甲虫iDNA技术,用于哺乳动物多样性监测和生态学研究
Abstract
enMany species of dung beetles (Coleoptera: Scarabaeidae: Scarabaeinae) are coprophages and possess trophic connections with mammalian dung. Recent studies have shown that the use of genetic information from dung beetle guts (invertebrate-derived DNA or iDNA) allows for the detection of mammals in a given habitat without intensively surveying the area. However, these studies used live or freshly killed beetles instead of preserved specimens. Here, we assessed the feasibility of extracting and sequencing iDNA from dung beetles that were collected using conventional baited pitfall traps with a mixture of propylene glycol and ethanol. We extracted iDNA from the guts of 18 dung beetles, comprising three species and three functional groups, collected from a seasonal tropical forest in Xishuangbanna, China. Eight mammalian species were detected, including rare species not previously recorded at the same location. Among the three functional groups, paracoprids (tunnelers) yielded the highest number of mammal species. Our study shows that iDNA can be successfully sequenced from preserved specimens, provided they are stored under appropriate conditions. The proposed technique offers a viable alternative to traditional cafeteria experiments for understanding dung beetle-mammal interactions and can serve as a valuable complementary approach to current mammal survey techniques.
摘要
zh许多粪甲虫物种(Coleoptera: Scarabaeidae: Scarabaeinae)以哺乳动物的粪便作为食物来源,因此与其存在营养层级上的关联。最近的研究表明,利用粪甲虫肠道内的遗传信息(取自无脊椎动物的DNA或iDNA),可检测出生活在特定栖息地的哺乳动物的种类,而无需直接对哺乳动物展开广泛调查。然而,这些研究使用的是活的或刚杀死的粪甲虫,而非粪甲虫标本。本研究评估了从常规陷阱诱捕、保存在丙二醇和乙醇混合物中的粪甲虫标本中提取iDNA并测序的可行性。研究使用的18只粪甲虫标本来自中国西双版纳季节性热带森林,包括来自3个功能类群的3个物种。结果共检测出8种哺乳动物,包括未在该地点记录过的稀有物种。在这3个功能群中,从外生型粪甲虫(隧道者)中检测到的哺乳动物种类最多。本研究表明,只要粪甲虫标本保存得当,可以从其肠道中提取iDNA并测序。本研究使用的技术可替代传统的自助餐式食物选择实验,用于了解粪甲虫与哺乳动物的相互作用,是目前哺乳动物调查技术的重要补充。【审阅:范欢】
Plain language summary
enMany dung beetles utilize mammalian dung as a food source and build and relocate dung balls to lay eggs and shelter their larvae. Such behavior provides many ecosystem services such as nutrient cycling, secondary seed dispersal, soil excavation, and parasite and pest control. Their relationships with mammal dung helped us detect which mammals are present in an area by looking at the DNA inside dung beetle guts. In the past, they used live or freshly killed dung beetles for these DNA studies. However, in this study, we have tested if we can use preserved dung beetles, such as those caught in pitfall traps with alcohol-based preservative solution, to obtain the same information. We successfully extracted DNA from the preserved beetles and used it to identify nonhuman mammal species. We even found rare mammal species that had not been spotted in that area before. Our study showed that we do not always need live beetles to extract mammal DNA. Preserved beetles can work, too, as long as they are stored correctly. This method helps scientists discover which mammals are in an area, and it is more efficient than other methods.
通俗语言摘要
zh许多粪甲虫物种利用哺乳动物的粪便作为食物,将其做成粪球,并在上面产卵。为保护幼虫,它们还会把粪球推到安全的地方。这些行为提供了许多生态系统服务,如营养循环、二次种子传播、松土,以及寄生虫与害虫控制。因为这种取食关系,我们可以通过检测粪甲虫肠道内的DNA,来判断粪甲虫活动区域内有哪些哺乳动物存在。在过去,研究人员使用活的或刚杀死的粪甲虫进行研究。本研究则测试了从粪甲虫标本中获取这些信息的可行性。这些标本虽然曾浸泡在酒精类保存溶液中,但仍可成功地从它们的肠道中提取到DNA,并检测出这些DNA来自哪些哺乳动物。我们甚至发现了以前在该地区未报道过的稀有哺乳动物。研究结果表明,并不总是需要从活的粪甲虫中提取哺乳动物的DNA,保存较好的粪甲虫标本也可以做此用途。这种方法可帮助研究人员监测某个区域生活着哪些哺乳动物。
Practitioner points
en
-
Dung beetles collected by conventional pitfall traps, which preserve specimens with an alcohol-based solution, can be used to extract, sequence, and identify mammal DNA from their guts.
-
This method identified a previously unrecorded rare Asian black bear, primarily nocturnal masked palm civet and northern pig-tailed macaque, demonstrating its potential to capture mammals across various spatiotemporal residencies.
-
The proposed technique offers a viable alternative to traditional cafeteria experiments for understanding dung beetle-mammal interactions and can serve as a valuable complementary approach to current mammal survey techniques.
实践者要点
zh
-
可从传统陷阱诱捕方法收集并保存在酒精类容液中的粪甲虫标本肠道中提取DNA并测序,检测到哺乳动物DNA。
-
本研究利用该技术,检测出夜间活动的果子狸、北方猪尾猕猴,以及一种以前没有记录过的稀有亚洲黑熊,证明该技术可检测到大尺度时空内的哺乳动物。
-
该技术可替代传统的自助餐式食物选择实验,用于了解粪甲虫与哺乳动物的相互作用,是目前哺乳动物监测技术的重要补充。
1 INTRODUCTION
Understanding interspecific interactions is important to assess the specific roles of species in complex ecological networks (Tylianakis & Morris, 2017) and to develop effective strategies for biodiversity conservation (Genes et al., 2017). Recent advances in molecular techniques have significantly improved our knowledge of ecological networks (Evans et al., 2016). Molecular-based techniques, such as metabarcoding of insects or their intestinal contents (invertebrate-derived DNA [iDNA]), have emerged as a subset of environmental DNA (eDNA) applications (Massey et al., 2022), allowing us to identify and quantify the strength of ecological interactions within trophic networks (Ji et al., 2022).
Many dung beetles (Coleoptera: Scarabaeidae: Scarabaeinae) are coprophages with obligatory trophic associations with mammalian dung (Bogoni et al., 2019; Halffter & Halffter, 2009). Adult dung beetles utilize mammalian dung as a food source and build and relocate brood balls to lay eggs and shelter their larvae (Scholtz et al., 2009). Dung beetles can be classified into three functional groups, namely telecoprids (rollers), paracoprids (tunnelers), and endocoprids (dwellers), based on their dung-relocation behavior (Tonelli, 2021). Telecoprids relocate dung and bury it away from the original dung pad. Paracoprids excavate tunnels beneath dung pads. Endocoprids reside within the dung pad itself. The relocation and consumption of dung by functionally variable beetles assist ecological processes and services such as nutrient cycling, secondary seed dispersal, soil excavation, and parasite and pest control (Nichols et al., 2008).
Gillett et al. (2016) used the iDNA approach to extract mammalian DNA from dung beetle guts and demonstrated the potential to employ molecular techniques to explore dung beetle–mammal interactions. The methodological details of the iDNA approach on dung beetles were also explored by Gómez and Kolokotronis (2017), who used Sanger sequencing to show that the detection probability of mammalian DNA depends on beetle body size. Subsequently, Drinkwater et al. (2021) demonstrated that the use of iDNA on dung beetles can serve as a temporal window to recover iDNA from their mammalian diet, providing a tool to monitor mammalian communities. These studies, however, used live beetles. Live beetles captured by manual hand collection may be taxonomically biased, as this method generally captures endocoprids from dung pads (Mora-Aguilar et al., 2023). On the other hand, pitfall trapping of live beetles can capture different beetle species and functional groups; however, captured live specimens have a higher chance of losing iDNA due to excretion.
The application of the iDNA approach on dung beetles has great potential if mammalian DNA can be detected from specimens collected by conventional pitfall traps. Conventional pitfall traps consist of a bait—derived from human or other mammalian dung, or alternative potential food sources for dung beetles (e.g., rotting fruits and mushrooms), and a collection pot with a killing and preservative solution (e.g., ethanol, propylene glycol, and water). Traps are left in the field for 24–48 h, depending on the spacing between traps and other environmental conditions (Larsen & Forsyth, 2005; Silva & Hernández, 2015). Ethanol effectively preserves collected specimens; however, pitfall traps with only ethanol solution cannot be left in the field for an extended duration due to its quick evaporation (Aristophanous, 2010). Consequently, a 75% propylene glycol and 25% ethanol solution is commonly used, as this can be kept in the field longer (Scholtz et al., 2009). The effectiveness of the iDNA approach hinges on the possibility of extracting mammalian DNA from dung beetles captured and preserved by such a method, but this has yet to be investigated.
This study aims to demonstrate the practicality of using baited pitfall traps with the conventionally used preservative solution to extract mammalian DNA from dung beetle guts. Such an approach not only offers insights into dung beetle–mammal interactions but also introduces an additional method for monitoring mammalian communities.
2 MATERIALS AND METHODS
2.1 Sample collection
Dung beetles were collected from a seasonal tropical forest in Bubeng, Xishuangbanna, China, as part of the dung beetle community assessment project in the Forest Global Earth Observatory 20-hectare forest dynamics plot (FDP) (Davies et al., 2021). The study site experiences an average annual rainfall of 1500 mm and a mean temperature of 18.5°C, characterizing a tropical monsoon climate (Cao et al., 2006). Sampling was conducted in August 2021, during the rainy season, across five one-hectare plots (100 × 100 m) established at the four corners and the center of the FDP (Supporting Information: Figure S1a). At the center of each one-hectare plot, we deployed a single pitfall trap baited with ~25 g of human dung. Each trap's collection cup was filled with a preservative solution consisting of 200 mL of propylene glycol and 50 mL of 99% ethanol (Supporting Information: Figure S1b). Traps were left in the plot for 48 h. The collected traps were transferred to 99% ethanol and stored at −20°C.
2.2 Dissection and gut extraction
We selected one species of dung beetle from each of the three functional groups for gut extraction: Onthophagus diabolicus (paracoprid), Onthophagus cf. gracilipes (endocoprid), and Paragymnopleurus sp. (telecoprid). The functional group of each species was determined based on a combination of information from published literature (Espinoza & Noriega, 2018), morphological characteristics, field observations, and consultations with local taxonomists (Nimalrathna et al., unpublished data). Before processing each specimen, all dissection tools were sterilized using an autoclave, rinsed in 95% alcohol, and exposed to a Bunsen burner flame until they glowed red. Distilled water, Eppendorf tubes, forceps, and needles were autoclaved at 121°C for 20 min. To minimize contamination risks, we employed a three-step procedure for extracting the gut of dung beetles (Gómez & Kolokotronis, 2017). First, the beetles' exoskeletons were thoroughly rinsed with distilled water, then with 95% v/v ethanol, and again with distilled water to remove adhesion particles. The elytra, wings, and abdominal tergites were then removed using forceps and a syringe needle. Finally, the abdomen contents were scooped out using a syringe needle and stored in Eppendorf tubes with 99% v/v alcohol at −20°C.
2.3 DNA extraction, amplification, and sequencing of dung beetle guts
Total DNA from the dung beetle guts was extracted using Zymo Research BIOMICS DNA Microprep Kit (Cat# D4301) following the manufacturer's instructions. Due to the possible degradation of mammalian DNA in dung beetles, we amplified a short segment of the mammalian mitochondrial 16S gene (60–84 bp) using MamP007F,5′-CGAGAAGACCCTATGGAGCT-3′ and Mam007R, 5′–CCGAGGTCRCCCCAACC–3′ primers (Giguet-Covex et al., 2014). We performed PCR amplification using the Applied Biosystems® PCR System 9700. The amplification of the samples produced a sequence library, which was then subjected to high-throughput sequencing on the PCR product using the Illumina Novaseq sequencing platform (see Supporting Information for more details).
2.4 Sequence processing, mammal identification, and composition
DNA sequences were analyzed through the DADA2 pipeline in R (Callahan et al., 2016). Sequences with low-quality ends were first trimmed to a length of 100 bp. Then, amplicon sequence variants (ASVs) were identified in each sample with chimeras removed. Finally, the ASVs were classified using the assignTaxonomy function in the DADA2 package, benchmarked against a customized reference database described as follows.
We collected information on mammalian species (except bats) occurrences in Xishuangbanna to build the reference database from various sources, including published literature (Supporting Information: Table S1), unpublished camera trap data from the study site, and a local checklist of mammals maintained by the environmental education center at Xishuangbanna Tropical Botanical Garden. Then, we obtained the mitochondrial genome data for each listed mammalian species using the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov) (see Supporting Information: Table S2 for the accession numbers used for each mammalian species). Geneious Prime software 2022.2.2 (https://www.geneious.com) was used to extract the 16S mitochondrial gene from the complete mitochondrial genome.
2.5 Data processing and visualization
We used R 4.2.1 (R Core Team, 2022) for all data analyses and illustrations described below. Given the exploratory nature of our study and its limited sample size, we have chosen to present the results descriptively. We first used the plot_alpha_rarefaction function in the microbiomeutilities package (Shetty, 2019) to generate a rarefaction curve, which allowed us to assess the sequencing depth specific to each sample. Sequencing depth refers to the overall count of sequences acquired from individual samples. It is essential to detect rare or low-abundance species with higher accuracy, thus enhancing the overall reliability of the assessment. We utilized the plot_composition function in the microbiome package (Lahti & Shetty, 2017) to visualize the composition of nonhuman mammalian DNA sequences in the dung beetle guts. To this end, we employed the tax_glom function to aggregate ASVs into species level and eliminated human DNA using the prune_taxa function in the phyloseq package (McMurdie & Holmes, 2013).
3 RESULTS
3.1 Sequencing summary and mammalian DNA identification in the dung beetle gut
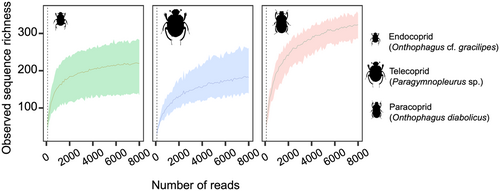
The study included 18 dung beetle specimens captured using pitfall traps. These specimens represent three species (paracoprid Onthopagus diabolicus, endocoprid Onthopagus cf. gracilipes, and telecoprid Paragymnopleurus sp.). However, from five beetles (two Onthopagus cf. gracilipes and three Paragymnopleurus sp.), nonhuman mammalian DNA could not be extracted, resulting in a recovery rate of 72%. After filtering and removing low-quality reads from the remaining successfully processed gut samples, we retrieved 744,688 reads. The number of retained sequences per sample varied between 37,184 and 48,153, averaging 41,372 reads per specimen. The rarefaction curves indicated that the paracoprids exhibited a greater sequence depth than the endocoprids and telecoprids (Figure 1). The rarefaction curve of paracoprids showed a steeper slope than those of endocoprids and telecoprids, but all curves reached asymptotes.
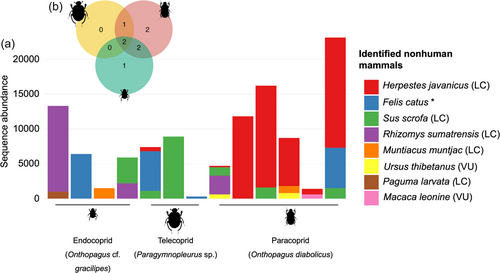
Based on published literature and other sources, we listed a total of 39 mammal species (excluding bats) known to occur in Xishuangbanna (Supporting Information: Table S2). We recovered eight nonhuman mammal species from dung beetle guts (Figure 2a). The identified mammals represented four orders and eight families, including common species (wild boar: Sus scrofa and the common muntjac: Muntiacus muntjac) and rare species (Asian black bear: Ursus thibetanus) known to occur in the habitat. Paracoprids yielded the highest number of mammal species (seven out of eight), with two or more mammal species detected from many individual specimens (Figure 2b). In contrast, endocoprids and telecoprids yielded lower numbers of mammal species. Most individual specimens from these groups detected only one or two mammal species, and notably, five specimens did not yield any nonhuman mammal species at all. Two mammal species were found uniquely in paracoprids, and one species in endocoprids (Figure 2b).
4 DISCUSSION
Until our study, the possibility of extracting dung beetle iDNA from conventionally used baited pitfall traps was uncertain. The amplified sequences in our study identified eight nonhuman mammal species. The composition of the detected species differed among the three functional groups but shared locally common wild boar (Sus scrofa) and domestic cat (Felis catus). Paracoprids preserved the highest number of mammal species (seven out of eight total recorded), including rare species such as the Asian black bear (Ursus thibetanus), southern red muntjac (Muntiacus muntjac), and the northern pig-tailed macaque (Macaca leonine). Telecoprids preserved the least mammal species, perhaps because they tend to rely on a single dung source and make brood balls from large and nutritious dung pads often derived from large omnivores (e.g., wild boars) (Batilani-Filho & Hernandez, 2017; Carpaneto et al., 2005). On the contrary, paracoprids represented diverse mammal diets, including those detected by telecoprids. Paracoprids' diverse diet may be related to their feeding preference for different types of mammal dung with highly variable dung sizes (Holter et al., 2002). The fact that endocoprids have a diet similar to both telecoprids and paracoprids is related to their primary behavior of feeding on smaller dung particles that have already been processed by telecoprids and paracoprids, which helps them minimize direct competition for resources (Holter et al., 2002; Sabu et al., 2006).
This approach provides an additional sampling tool for not only the discovery of mammals but also the studies of dung beetle–mammal interactions. We demonstrate that dung beetle specimens captured by conventionally used baited pitfall traps can preserve the DNA of mammal species in their guts. Conventionally used baited pitfall traps can capture diverse dung beetle species with various mammalian diets over an extended period in the field. As a result, it enables the detection of a broader range of mammalian species through iDNA compared to live beetles collected by hand. Notably, we detected the presence of the Asian black bear, which had not been recorded in previous camera trap studies specific to this location, although it has been listed in the greater Xishuangbanna region (Tongkok et al., 2020). Additionally, this approach detected two arboreal species, the masked palm civet (Paguma larvata) and the northern pig-tailed macaque (Macaca leonine), demonstrating that the beetles preserved DNA from a diverse array of mammals in their gut, encompassing common, rare, and even arboreal species. The conventional dung beetle–mammal interaction experiments, such as cafeteria experiments involving selected dung sources (Raine et al., 2018) and laboratory dietary trials (Jones et al., 2012), may overlook potential interactions with rare or niche-specialized mammal species. Nonetheless, the number of mammal species identified using dung beetle iDNA was lower than the number of species known to occur at the study site. The sequencing depth for each species reached asymptote, indicating that we obtained enough sequences to capture mammalian DNA. Consequently, the detected disparity between observed and listed mammalian species could be attributed to the small sample size, the specific choice of beetle species for investigation, or the possibility that some mammal species no longer inhabit the studied forest area. We expect that the inclusion of more dung beetle species would yield more mammal species, as our recent survey in the same region found over 50 species (Nimalrathna et al., unpublished data). Our methods do not replace other mammal monitoring methods (e.g., camera traps) but complement other tools for monitoring mammal populations.
Our study showed that dung beetles collected from conventionally used baited pitfall traps preserved mammal DNA in their guts, demonstrating the use of dung beetle iDNA as an additional tool for biodiversity assessment. Our study suggests that paracoprids are an ideal focal group to survey mammal species effectively. Paracoprids are commonly collected by baited pitfall traps (Mora-Aguilar et al., 2023), thus reinforcing our approach as an effective method to monitor mammals. Compared to live dung beetles, the use of preserved dung beetles captured by baited pitfall traps makes sampling easier and more cost-effective while minimizing sampling bias. This method may also enable us to detect mammal species that are already rare and threatened by anthropogenic disturbances (e.g., the Asian black bear). Similar to other iDNA applications designed to understand insect-plant (Kajtoch et al., 2015) and predator-prey (Cuff et al., 2023) interactions, the proposed approach broadens the capacity of the dung beetle community and trophic interaction studies.
AUTHOR CONTRIBUTIONS
Thilina S. Nimalrathna: Conceptualization; data curation; formal analysis; investigation; methodology; project administration; software; visualization; writing—original draft. Huan Fan: Methodology; software; validation; visualization; writing—review and editing. Rui-Chang Quan: Resources. Akihiro Nakamura: Funding acquisition; project administration; resources; supervision; validation; visualization; writing—review and editing.
ACKNOWLEDGMENTS
Thilina Nimalrathna expresses gratitude for the support provided by the Belt and Road Chinese Government Scholarship and The Alliance of International Science Organizations (ANSO) PhD scholarship. The research was primarily funded through grants from Akihiro Nakamura (the National Natural Science Foundation of China International [Regional] Cooperation and Exchange Project [32161160324], the 14th Five-Year Plan of the Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences [E3ZKFF1K], and the High-End Foreign Experts Program of the High-Level Talent Recruitment Plan of Yunnan Province) and Ahimsa Campos-Arceiz (Southeast Asia Biodiversity Research Institute, Grant/Award Number: Y4ZK111B01).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data will be made available on request.




