Colposcopy referrals and CIN3 detection after triage by host cell DNA methylation and/or HPV genotyping in HPV positive women with low-grade cytology from a population-based Dutch primary HPV screening trial
Abstract
High-risk HPV (hrHPV)-based screening has led to many unnecessary colposcopy referrals, mainly because of direct referral after low-grade cytology (ASC-US/LSIL). DNA methylation and genotyping tests on ASC-US/LSIL samples have the potential to significantly improve the efficiency of screening. In this study, 12 triage strategies were constructed from FAM19A4/miR124-2 or ASCL1/LHX8 methylation, HPV16/18 or HPV16/18/31/33/45 genotyping and 1-year repeat cytology. The performance was evaluated on 215 hrHPV-positive ASC-US/LSIL samples from the IMPROVE trial (NTR5078). Performance was measured by colposcopy referral rate, positive predictive value (PPV) for detecting precancer (CIN3), and negative predictive value (NPV). To evaluate efficiency, strategies were ordered by the cumulative colposcopy referral rate after 1-year cytology and compared by the marginal PPV to detect one additional CIN3 (mPPV). The most conservative strategy (referral when HPV16/18 and FAM19A4/miR124 methylation results are positive) had a direct referral rate of 5.2%, a cumulative referral rate after 1-year cytology of 54.1%, and mPPV of 19.3%. Replacing HPV16/18 by HPV16/18/31/33/45 increased the cumulative 1-year referral rate to 54.6%, and yielded an mPPV of 10.0%. Similar results were obtained for strategies with ASCL1/LHX8 methylation. Of all strategies, referral after an HPV16/18/31/33/45 positive, ASCL1/LHX8 methylation-positive, and/or 1-year cytology-positive result yielded the highest direct and cumulative 1-year colposcopy referral rates of 64.4% and 79.1%, respectively. The NPVs after 1-year cytology varied between 98.1% and 99.4%, warranting a return to routine screening. Altogether, DNA methylation-based triage strategies are recommended as they are discriminative for CIN3 and control the number of immediate colposcopy referrals.
What's New?
The implementation of primary human papillomavirus (HPV)-based cervical cancer screening has led to an increase in colposcopy referrals and detection of low-grade lesions. This study examines the ability of methylation-based triage strategies, together with HPV genotyping, to improve screening efficiency in women with low-grade cytology. Analyses show that the combination of HPV16/18 genotyping and FAM19A4/miR124-2 methylation resulted in a 95% reduction in direct colposcopy referrals. As single triage tests, FAM19A4/miR124-2 or ASCL1/LHX8 methylation decreased direct referrals by 86%. The findings indicate that methylation-based triage strategies help reduce colposcopy referrals and maintain timely detection of clinically relevant lesions.
1 INTRODUCTION
Human papillomavirus (HPV)-based cervical cancer screening has emerged as a highly sensitive method for detecting high-grade cervical intraepithelial neoplasia (CIN) and cervical cancer, surpassing the sensitivity of cytology-based screening.1, 2 However, it is important to address the issue of lower specificity, leading to a significant increase in colposcopy referrals.3, 4 Notably, a significant proportion of these referrals do not have clinically relevant disease (≤CIN1).5, 6 In hrHPV-positive women with low-grade cytology (atypical squamous cells of undetermined significance (ASC-US); or low-grade squamous intraepithelial lesions (LSIL)), the prevalence of CIN3 or cancer (CIN3+) is about 10% underlining the unmet need for improved triage strategies. In the Netherlands, additional HPV16/18 genotyping for ASC-US/LSIL samples was introduced in July 2022, with immediate referral after an HPV16/18-positive result and repeat testing after 1 year when a woman was non-HPV16/18-positive. This genotyping strategy aims to reduce unnecessary colposcopy referrals while ensuring early detection of clinically significant cases.
Understanding the molecular mechanisms involved in cervical carcinogenesis is crucial for optimising screening strategies. In this context, it is well established that hypermethylation of promoter regions of specific tumour suppressor genes plays a pivotal role in cervical carcinogenesis.7, 8 As the methylation levels of these tumour suppressor genes increase, the risk of progression of CIN lesions to cervical cancer also increases, with the highest methylation levels observed in women with cervical cancer.9-11 DNA methylation biomarkers have emerged as potential triage tools for HPV-based cervical cancer screening, exhibiting high sensitivity for cervical cancer and ‘advanced CIN lesions’, which refer to CIN2/3 lesions associated with an hrHPV infection lasting over 5 years.7, 10-13 The effectiveness of these triage markers has been evaluated in terms of clinical performance for CIN3+,12-18 the number of colposcopy referrals15, 19 and the safety of a negative test, particularly the minimal risk of progression to CIN3 or carcinoma following a negative methylation result.19, 20
Previous studies have demonstrated the potential of FAM19A4/mir124-2 methylation and ASCL1/LHX8 as additional risk stratification tools in hrHPV-positive women with ASC-US/LSIL.13, 15, 19 The study by Dick et al., which included participants from the population-based POBASCAM trial and the VUSA-screen trial, indicated that by incorporating FAM19A4/miR124-2 methylation testing, a substantial 58.8% reduction in direct colposcopy referrals can be achieved while maintaining a high sensitivity of 70.2% for detecting CIN3+.19 Similar results were reported by Bonde et al.,15 showing a 66% reduction in colposcopy referrals for hrHPV-positive women with ASC-US/LSIL cytology using FAM19A4/miR124-2 methylation testing. These findings underscore the potential of DNA methylation biomarkers in cervical cancer screening programs to improve the efficiency of screening by reducing colposcopy referral while retaining high sensitivity. While reducing colposcopy referrals is valuable, it is crucial to remember that high-grade CIN is heterogeneous. A negative methylation test indicates a low cancer risk.20
In light of these developments, our study aims to assess the effectiveness, particularly in terms of positive predictive value (PPV) and negative predictive value (NPV) of various molecular triage methods, including FAM19A4/miR124-2 methylation, ASCL1/LHX8 methylation, HPV16/18 genotyping, extended genotyping with HPV16/18/31/33/45, and combinations thereof, in hrHPV-positive women with ASC-US/LSIL cytology from a population-based Dutch primary HPV screening trial (IMPROVE study21).
2 METHODS
2.1 Clinical specimens
This study involved a post-hoc analysis of the IMPROVE trial (The Netherlands Trial Register NTR5078). The IMPROVE trial was designed as a randomised non-inferiority trial to assess the clinical accuracy of HPV testing on self-collected samples compared to clinician-collected cervical samples within the Dutch cervical cancer screening program. Approval for the IMPROVE trial was obtained from the Ministry of Public Health (The Hague, The Netherlands; IMPROVE VWS no. 2014/32). A total of 16,410 women participated in the trial and were randomly assigned in a 1:1 ratio to either the intervention group (self-sampling) or the control group (clinician-based sampling). All 1020 hrHPV-positive women underwent re-testing using the other collection method. Detailed information about the IMPROVE trial can be found in a previous publication by Polman et al.21 For this study, we included all 215 women with an hrHPV-positive clinician-collected cervical sample with an ASC-US/LSIL cytology result. According to the latest classifications of the World Health Organisation (WHO), histology was categorised into normal (no dysplasia; CIN0), LSIL/CIN1, HSIL/CIN2, and HSIL/CIN3, which are hereafter referred to as CIN1, CIN2, and CIN3, respectively.22 Follow-up data was collected for all women. All participants provided informed consent for additional follow-up research.
2.2 HPV genotyping, FAM19A4/miR124-2 methylation and ASCL1/LHX8 methylation
HPV DNA testing was performed using the hrHPV GP5+/6+ PCR enzyme immunoassay (EIA; Labo Biomedical Products BV, Rijswijk, The Netherlands). This assay detects 14 high-risk HPV types, that is, HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68. GP5+/6+ PCR amplicons were genotyped using the LMNX Genotyping kit HPV GP HR (Diassay, QIAgen, Hilden, Germany) according to the instructions of the manufacturer.
Methylation analysis of ASCL1/LHX8 was conducted as previously described13, 16, 23 using quantitative methylation-specific PCR on bisulfite-converted DNA. The ASCL1/LHX8 methylation status was labelled positive if the result exceeded the pre-defined predicted probability threshold.12 FAM19A4/miR124-2 methylation analysis was performed using the QIAsure Methylation Test® (Qiagen, Hilden, Germany) on the same bisulphite-converted DNA samples according to the instructions of the manufacturer. The FAM19A4/miR124-2 methylation status was labelled positive if the result surpassed the predefined ΔΔCq value threshold, in accordance with the manufacturer's instructions. Only samples with a valid test result for all assays were included in this study.
2.3 Data and statistical analysis
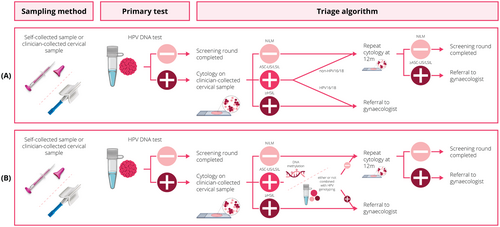
The current study evaluated the performance of 12 single and combined triage strategies utilising FAM19A4/miR124-2 methylation, ASCL1/LHX8 methylation, HPV16/18 genotyping and/or HPV16/18/31/33/45 genotyping. A triage result was labelled positive when test positivity was shown for (I) FAM19A4/miR124-2 methylation, (II) ASCL1/LHX8 methylation, (III) HPV16/18 genotyping, (IV) HPV16/18/31/33/45 genotyping, (V) HPV16/18 genotyping AND FAM19A4/miR124-2 methylation, (VI) HPV16/18 genotyping OR FAM19A4/miR124-2 methylation, (VII) HPV16/18 genotyping AND ASCL1/LHX8 methylation, (VIII) HPV16/18 genotyping OR ASCL1/LHX8 methylation, (IX) HPV16/18/31/33/45 genotyping AND FAM19A4/miR124-2 methylation, (X) HPV16/18/31/33/45 genotyping OR FAM19A4/miR124-2 methylation, (XI) HPV16/18/31/33/45 genotyping AND ASCL1/LHX8 methylation and (XII) HPV16/18/31/33/45 genotyping OR ASCL1/LHX8 methylation.
Direct colposcopy referral rate, PPV, and NPV for detection of CIN3 were estimated together with Wald 95% confidence intervals (95%CI). The direct referral percentage was calculated as the percentage of triage positives. The cumulative colposcopy referral rate and NPV after 1-year cytology testing were adjusted for loss to follow-up and the imperfect sensitivity of cytology. The sensitivity of cytology was estimated from the POBASCAM population-based HPV screening study25, 29, 35 in which women with AS-CUS/LSIL were repeated twice by cytology or combined HPV testing and cytology. The estimated sensitivity of repeat cytology after AS-CUS/LSIL was 90% (47/52, 95%CI 0.82–0.98) for detection of CIN3+ and 85% (79/93, 95%CI 0.78–0.92) for CIN2+. Jeffreys 95%CIs were used for the 1-year referral percentage and NPV after repeat testing, accounting for uncertainty in the estimated sensitivities.
To compare the efficiency of the different strategies, we utilised the incremental cost-effectiveness (ICER) framework originally developed for health technology assessments.24, 25 In our situation, costs are set equal to the number of cumulative colposcopy referrals after 1-year repeat cytology, and effects are set equal to the number of detected CIN3. To identify strategies that strike an optimal balance between costs and effects in a cost-effectiveness study, adjacent non-dominated strategies are compared by means of the ICER. In our setting, a strategy is called non-dominated when there does not exist another strategy or combination of two other strategies that yields a higher number of CIN3 against the same number of referrals. An insightful measure is the inverse of the ICER or marginal PPV (mPPV), which is the additional number of CIN3 detected per additional colposcopy referral. The curve connecting adjacent non-dominated strategies in a two-dimensional plot of number of colposcopy referrals against number of CIN3 is the efficient frontier. To select strategies from the efficient frontier, we used a Dutch PPV threshold of 20%,26 which is based on the PPV of abnormal cytology for CIN3+ in the previous cytology-based program until year 2016 which was 20%–30%.27 To evaluate whether a negative triage warrants return to routine screening, we used the Dutch NPV threshold of 98%, which is based on the CIN3 risk after negative cytology-based screening program.26
All statistical analyses were performed with SPSS Statistics (version 28, IBM Corp, Armonk, NY, USA).
3 RESULTS
A total of 215 hrHPV-positive women with ASC-US/LSIL cytology were identified in the IMPROVE trial and were available for HPV genotyping and methylation analysis. Six samples (6/215; 2.3%) with invalid methylation results were excluded from further analysis. Additionally, 15 samples (15/215; 7.0%) lacked genotyping results and were also excluded from further analysis, resulting in a total of 194 hrHPV-positive women for the final analysis. Among these, 22 (11.3%) were diagnosed with CIN3, 30 (15.5%) with CIN2 and 142 (73.2%) had no evidence of CIN2+, including 67 with CIN1 and 52 CIN0 and 23 women with two consecutive normal cytology results. The median age of the women included in the study was 39 years (range: 29–60 years). Out of the 194 women, 28 (14.4%) women tested positive for FAM19A4/miR124-2 methylation, 57 (29.4%) women tested positive for ASCL1/LHX8 methylation, 71 (36.6%) women tested positive for HPV16/18, and 105 (54.1%) women tested positive for HPV16/18/31/33/45. Among the 142 women without CIN2/3, 90 women (63.4%) underwent cytology screening after 1-year. Among these, 52 (57.8%) had normal cytology, 36 (40.0%) ASC-US/LSIL cytology and 2 (2.2%) HSIL cytology.
Table 1 presents the PPV and NPV for all single and combined triage strategies for detecting CIN3 (Supplementary Table 1 for CIN2+). Triage strategies are highlighted in bold if there is no other strategy with equal or higher values for both PPV and NPV. The strategies presented vary widely with respect to the direct colposcopy referral rate. However, the difference between the colposcopy referral rates becomes smaller after 1-year repeat cytology. HPV16/18 genotyping AND FAM19A4/miR124-2 methylation (strategy V) is the most conservative strategy with a direct colposcopy referral rate of 5.2% (95%CI 2.0%–8.3%), and a PPV for CIN3 of 50.0% (95%CI 19.0%–81.0%). The cumulative colposcopy referral rate after 1-year is 54.1% (95%CI 47.4%–61.3%). A similar strategy is HPV16/18/31/33/45 genotyping AND FAM19A4/miR124-2 methylation (strategy IX) with a direct colposcopy referral rate of 6.7% (95%CI 3.2%–10.2%), PPV for CIN3 of 46.2% (95%CI 19.1%–73.3%) and 1-year colposcopy referral rate of 54.6% (95%CI 47.9%–61.2%). The most aggressive strategy, including HPV16/18/31/33/45 genotyping OR ASCL1/LHX8 methylation (strategy XII), has a direct colposcopy referral rate of 64.4% (95%CI 57.7R–71.2%), and achieves a PPV for CIN3 of 15.2% (95%CI 8.9%–21.5%). The colposcopy referral rate increases to 79.1% after 1 year (95%CI 72.9%–84.3%) No strategy meets the 98% NPV threshold at baseline, indicating that none of the strategies warrants return to routine screening. However, all strategies achieve a 98% NPV after 1-year repeat cytology (range 98.1%–99.4%).
| PPV | NPV | Colposcopy referral | NPV (after 1-year repeat cytology) | Colposcopy referral (with 1-year repeat cytology) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Triage strategy | % | 95%CI | % | 95%CI | % | 95%CI | % | 95%CI | % | 95%CI | |
| I | FAM19A4/miR124-2 methylation | 32.1% | 14.8%–49.4% | 92.2% | 88.1%–96.3% | 14.4% | 9.5%–19.4% | 98.5% | 94.0%–99.8% | 58.4% | 51.6%–65.4% |
| II | ASCL1/LHX8 methylation | 24.6% | 13.4%–35.7% | 94.2% | 90.2%–98.1% | 29.4% | 23.0%–35.8% | 98.9% | 94.3%–99.9% | 63.5% | 56.7%–70.2% |
| III | HPV16/18 genotyping | 18.3% | 9.3%–27.3% | 92.7% | 88.1%–97.3% | 36.6% | 29.8%–43.4% | 98.7% | 93.6%–99.9% | 66.2% | 59.4%–72.7% |
| IV | HPV16/18/31/33/45 genotyping | 14.3% | 7.6%–21.0% | 92.1% | 86.5%–97.7% | 54.1% | 47.1%–61.1% | 98.6% | 92.1%–99.9% | 75.3% | 69.1%–81.1% |
| V | HPV16/18 genotyping AND FAM19A4/miR124-2 methylation | 50.0% | 19.0%–81.0% | 90.8% | 86.6%–94.9% | 5.2% | 2.0%–8.3% | 98.2% | 93.9%–99.7% | 54.2% | 47.4%–61.3% |
| VI | HPV16/18 genotyping OR FAM19A4/miR124-2 methylation | 19.1% | 10.9%–27.3% | 95.2% | 91.2%–99.3% | 45.9% | 38.9%–52.9% | 99.2% | 93.9%–100.0% | 70.4% | 63.9%–76.6% |
| VII | HPV16/18 genotyping AND ASCL1/LHX8 methylation | 33.3% | 14.5%–52.2% | 91.8% | 87.6%–95.9% | 12.4% | 7.7%–17.0% | 98.4% | 93.9%–99.8% | 57.2% | 50.4%–64.2% |
| VIII | HPV16/18 genotyping OR ASCL1/LHX8 methylation | 18.3% | 10.8%–25.7% | 96.7% | 93.0%–100.4% | 53.6% | 46.6%–60.6% | 99.5% | 94.2%–100.0% | 72.7% | 66.3%–78.7% |
| IX | HPV16/18/31/33/45 genotyping AND FAM19A4/miR124-2 methylation | 46.2% | 19.1%–73.3% | 91.2% | 87.0%–95.3% | 6.7% | 3.2%–10.2% | 98.2% | 93.9%–99.8% | 54.7% | 47.9%–61.8% |
| X | HPV16/18/31/33/45 genotyping OR FAM19A4/miR124-2 methylation | 14.9% | 8.5%–21.2% | 94.5% | 89.3%–99.7% | 61.9% | 55.0%–68.7% | 99.0% | 91.9%–100.0% | 79.1% | 73.1%–84.5% |
| XI | HPV16/18/31/33/45 genotyping AND ASCL1/LHX8 methylation | 27.0% | 12.7%–41.3% | 92.4% | 88.2%–96.5% | 19.1% | 13.5%–24.6% | 98.5% | 94.0%–99.8% | 60.0% | 53.3%–67.0% |
| XII | HPV16/18/31/33/45 genotyping OR ASCL1/LHX8 methylation | 15.2% | 8.9%–21.5% | 95.7% | 90.8%–100.5% | 64.4% | 57.7%–71.2% | 99.3% | 92.5%–100.0% | 79.0% | 72.9%–84.3% |
- Note: A strategy is depicted in bold if there is no other strategy with a higher PPV and NPV.
When comparing methylation testing with HPV genotyping, methylation demonstrates superior discrimination in terms of both NPV and PPV. This can also be illustrated by looking at the CIN3 risk of combinations of methylation testing and HPV161/18 genotyping. The CIN3 risk after an HPV16/18-negative, FAM19A4/miR124-2-positive result is 22.2% (95%CI 6.4%–47.6%) and the CIN3 risk after an HPV16/18-negative, ASCL1/LHX8-positive result is 18.2% (95%CI 7.0%–35.5%). The CIN3 risks after a genotype positive, methylation negative result are markedly lower with a CIN3 risk of 13.1% (95%CI 5.8%–24.2%) after a HPV16/18-positive, FAM19A4/miR124-2-negative result and 10.6% (95%CI 3.6%–23.1%) after a HPV16/18-positive, ASCL1/LHX8-negative result. The latter risks are similar to the average CIN3 risk of 11.3% (95%CI 7.%–16.7%) in women with ASC-US/LSIL.
The incremental analysis is displayed in Figure 1. HPV16/18 genotyping (strategy III), HPV16/18/31/33/45 genotyping (strategy IV), HPV16/18/31/33/45 genotyping OR FAM19A4/miR124-2 methylation (strategy X), and HPV16/18/31/33/45 genotyping OR ASCL1/LHX8 methylation (strategy XII) are dominated because they lie clearly below the efficient frontier. Strategies on the efficiency frontier are HPV16/18 genotyping AND FAM19A4/miR124-2 methylation (strategy V), HPV16/18/31/33/45 genotyping AND FAM19A4/miR124-2 methylation (strategy IX), ASCL1/LHX8 methylation (strategy II), and HPV16/18 genotyping OR ASCL1/LHX8 methylation (strategy VIII) with mPPV values varying from 2.8% to 19.3%. Only strategy V has an mPPV close to the Dutch PPV threshold of 20%, although strategy V and IX perform very similarly in terms of cumulative colposcopy referral rate and number of CIN3+ detected.

4 DISCUSSION
The implementation of HPV-based screening in the Netherlands has led to a two-fold increase in direct colposcopy referrals and CIN1 diagnoses.5, 6, 28 Therefore, limiting the number of colposcopy referrals is pivotal when assessing triage strategies. Among the triage strategies, HPV16/18 genotyping AND FAM19A4/miR124-2 methylation (strategy V) had the lowest colposcopy referral rate and performed well in the incremental analysis with values of mPPV around the Dutch threshold of 20%. In terms of colposcopy referral rates, this strategy led to a 95% reduction of direct colposcopies compared to referring all women and 46% reduction in colposcopy referrals after 1-year cytology. Other strategies had a lower mPPV than the Dutch threshold, but HPV16/18/31/33/45 genotyping AND FAM19A4/miR124-2 methylation (strategy IX) may also be considered as it has a similar PPV and 1-year colposcopy referral rate as strategy V. When using FAM19A4/miR124-2 (or ASCL1/LHX8) methylation as a single triage test, the reduction in direct colposcopy referrals was 86% (or 71%) and the reduction in colposcopy referrals after 1-year was 42% (or 36%). The reduction in direct colposcopy referrals is in accordance with Dick et al. and Bonde et al., who reported reductions of 60% and 66% in colposcopy referral rates, respectively.15, 19
Previous research underlines the importance of secondary triage for hrHPV-positive women with ASC-US/LSIL cytology in a retrospective, co-testing19 and cross-sectional study.15 Our study builds upon these findings, capitalising on its distinct strength, and using samples from the prospective IMPROVE trial in Dutch primary HPV-based screening, aligned with the national protocol and offers an important population-based perspective. Including methylation as a triage test offers several advantages. First, methylation provides an objective molecular test to identify clinically relevant high-grade lesions. Second, a methylation test leads to more efficient utilisation of healthcare resources by significantly reducing the colposcopy referral rate. Third, our approach facilitates risk stratification, by reducing direct colposcopy referrals based on a low risk of CIN3, measured through the mPPV. It is essential to note that none of the strategies achieved an NPV of >98% after baseline triage, which has been defined as an informal threshold for return to routine screening in cervical cancer screening in The Netherlands.26 However, all estimated NPVs were the 98% NPV threshold26 after repeat cytology testing, to capture potentially missed CIN3 lesions. Previous studies of triage strategies in HPV-based screening also showed that strategies with repeat cytology, as currently implemented in the Netherlands, reach an NPV of at least 98%.29, 30 This supports the notion that immediate referral should be recommended only to those at highest CIN3 risk, and offering repeat testing otherwise. The methylation tests FAM19A4/miR124-2 and ASCL1/LHX8 methylation has demonstrated good sensitivity for cervical cancer and advanced CIN2/3 lesions, indicating the potential to identify lesions at highest risk of progression to cervical cancer.10-13, 23 This is further supported in longitudinal clinical studies where the absence of FAM19A4/miR124-2 methylation is associated with a high likelihood of spontaneous regression of CIN2/3 lesions31, 32 In addition, in longitudinal analyses of hrHPV-positive women, a lower 14-year cervical cancer risk was observed after negative FAM19A4/miR124-2 methylation test than after negative cytology.20 Taken together, in the context of cervical cancer screening and triage testing of hrHPV-positive women, the performance of FAM19A4/miR1242 and ASCL1/LHX8 methylation holds promise for identifying high-risk lesions, aiding in appropriate clinical management and minimising unnecessary invasive procedures for lesions likely to regress spontaneously.
A limitation of our study that the sensitivities of repeat cytology after ASC-US/LSIL, needed for the calculation of the NPV after repeat testing, were retrieved from the POBASCAM study.35 In this study, women with AS-CUS/LSIL were not immediately referred but followed by cytology or combined HPV testing and cytology. The estimated sensitivities of repeat cytology after ASC-US/LSIL for detection of CIN3+ and CIN2+ were high (90% and 85%, respectively), but are similar to the baseline sensitivities of cytology among HPV-positive women in our cohort of 89% and 85% for CIN3+ and CIN2+, respectively.13 Furthermore, the POBASCAM study and the current IMPROVE study are very similar, both are Dutch population-based HPV screening studies, and they both use the GP5+/6+ PCR as HPV screening test.35 Therefore, we think that the use of external data for the estimation of the sensitivity of cytology is warranted. Another limitation is the relatively modest number of CIN3 cases in our study. However, the CIN3+ risk in our cohort (11%, 22/194) is similar to a 9% CIN3+ risk observed after ASC-US/LSIL in the Dutch screening program,28 and the NPVs in our study were similar to those in the POBASCAM study where the NPV after negative repeat cytology for CIN3+ was 98.3%.25
The IMPROVE study was set up by Polman et al., and they reported an HPV prevalence of 7.3%.21 In a recent study by Inturissi et al., an increase in the HPV prevalence was observed, reaching nearly 10%.33 This notable increase in HPV prevalence highlights the mounting importance of implementing a conservative approach, particularly in the management of HPV-positive women with ASC-US/LSIL cytology. Such a conservative approach with DNA methylation analysis is also particularly important in the post-vaccination era, as methylation analysis targets the most clinically relevant lesions, which is crucial when evaluating lesions negative for HPV16/18.34
When assessing the full implications of a triage strategy, it is also important to consider the potential costs associated with incorporating a methylation test in comparison to HPV16/18 genotyping. While introducing the methylation test may entail upfront expenses, it is essential to recognise the downstream benefits of reducing unnecessary colposcopy referrals. This reduction in colposcopy referrals driven by improved risk stratification, has the potential to counterbalance the initial testing costs. Therefore, a comprehensive cost-effectiveness analysis should not only consider the direct testing expenses but also encompass repeat testing after 12 months and additional diagnostic tests (Figure 2).
In summary, our study demonstrates the potential of incorporating methylation-based strategies, specifically FAM19A4/miR124-2 and ASCL1/LHX8 to significantly reduce the number of colposcopy referrals, thereby improving the efficiency of cervical cancer screening. However, it is crucial to acknowledge that reducing colposcopy referrals also raises concerns about the potential risk of missing high-grade CIN lesions. Therefore, it is crucial to balance between efficiency and effectiveness, ensuring a comprehensive screening surveillance strategy to minimise the risk of missed high-grade CIN lesions.
AUTHOR CONTRIBUTIONS
Lisanne Verhoef: Data curation; formal analysis; methodology; validation; visualization; writing – original draft; writing – review and editing. Maaike C. G. Bleeker: Conceptualization; supervision; writing – review and editing. Nicole Polman: Data curation; resources; writing – review and editing. Kelsi R. Kroon: Visualization; writing – review and editing. Renske D. M. Steenbergen: Conceptualization; supervision; writing – review and editing. Renée M. F. Ebisch: Data curation; resources; writing – review and editing. Willem J. G. Melchers: Data curation; resources; writing – review and editing. Rudolf Lambertus Maria L.M. Bekkers: Data curation; resources; writing – review and editing. Anco C. Molijn: Data curation; resources; writing – review and editing. Folkert van Kemenade: Data curation; resources; writing – review and editing. Chris J. L. M. Meijer: Conceptualization; formal analysis; writing – review and editing. Daniëlle A. M. Heideman: Conceptualization; formal analysis; supervision; writing – review and editing. Hans Berkhof: Conceptualization; formal analysis; methodology; supervision; writing – review and editing.
ACKNOWLEDGEMENTS
We gratefully acknowledge late Wim Quint and Peter Snijders who were investigators of the IMPROVE study. We thank all women who participated in the IMPROVE study; the screening organisations Midden-West, Zuid-West, and Oost; the National Institute for Public Health and the Environment; and the physicians, gynaecologists, and pathologists in the study regions. The authors would like to acknowledge the nationwide network and registry of histo- and cytopathology in the Netherlands (PALGA) for providing data and the Amsterdam UMC Biobank for their high-quality storage of collected samples.
FUNDING INFORMATION
This project was funded by the Dutch Cancer Society (grant number KWF 11337) and the Horizon 2020 research and innovation program of the European Commission (RISCC project, grant agreement no. 847845).
CONFLICT OF INTEREST STATEMENT
DAMH, RDMS, and CJLMM are minority shareholders of Self-screen B.V., a spin-off company of VUmc (currently known as AmsterdamUMC location Vrije Universiteit Amsterdam); Self-screen B.V. develops, manufactures, and licenses high-risk HPV and methylation marker assays for cervical cancer screening and holds patents on these tests; CJLMM is part-time CEO of Self-screen B.V., and served occasionally on the scientific advisory boards of Qiagen; the other authors declare no conflicts of interests.
ETHICS STATEMENT
The work in this study was approved by the Medical Ethics Committee of Amsterdam UMC, Vrije Universiteit Amsterdam (Amsterdam, The Netherlands; METC 2018/09, TcB 2018.106). All participants provided written informed consent. The initial IMPROVE trial has been registered at the Netherlands Trial Register (NTR5078).
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of our study are in anonymised form available from the corresponding author upon reasonable request and following the data protection regulations.





