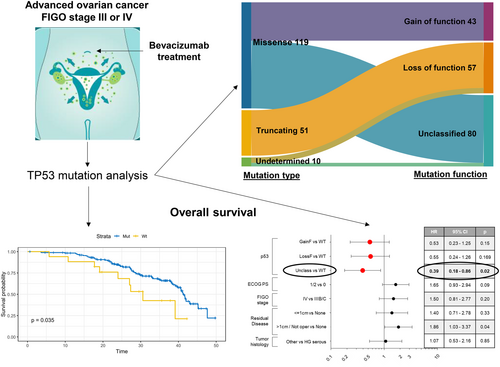
TP53 mutations and survival in ovarian carcinoma patients receiving first-line chemotherapy plus bevacizumab: Results of the MITO16A/MaNGO OV-2 study
Eliana Bignotti and Vittorio Simeon contributed equally to this study.
Abstract
To date, there are no biomarkers that define a patient subpopulation responsive to bevacizumab (BEV), an effective treatment option for advanced ovarian carcinoma (OC). In the context of the MITO16A/MaNGO OV-2 trial, a Phase IV study of chemotherapy combined with BEV in first-line treatment of advanced OC, we evaluated TP53 mutations by next-generation sequencing and p53 expression by immunohistochemistry (IHC) on 202 and 311 cases, respectively. We further correlated TP53 mutations in terms of type, function, and site, and IHC data with patients' clinicopathological characteristics and survival. TP53 missense mutations of unknown function (named unclassified) represented the majority of variants in our population (44.4%) and were associated with a significantly improved overall survival (OS) both in univariable (hazard ratio [HR] = 0.43, 95% confidence interval [CI] = 0.20–0.92, p = .03) and multivariable analysis (HR = 0.39, 95% CI = 0.18–0.86, p = .02). Concordance between TP53 mutational analysis and IHC was 91%. We observed an HR of 0.70 for OS in patients with p53 IHC overexpression compared to p53 wild-type, which however did not reach statistical significance (p = .31, 95% CI = 0.36–1.38). Our results indicate that the presence of unclassified TP53 mutations has favorable prognostic significance in patients with OC receiving upfront BEV plus chemotherapy. In particular, unclassified missense TP53 mutations characterize a subpopulation of patients with a significant survival advantage, independently of clinicopathological characteristics. Our findings warrant future investigations to confirm the prognostic impact of TP53 mutations in BEV-treated OC patients and deserve to be assessed for their potential predictive role in future randomized clinical studies.
What's New?
Bevacizumab (BEV) is an effective treatment for advanced ovarian cancer, but so far, there are no known biomarkers that indicate whether a patient will respond well to the drug. Here, the authors show that missense mutations in TP53 are associated with improved overall survival in patients receiving first-line chemotherapy plus BEV for advanced ovarian cancer. Based on these results, it may be useful in future studies to test patients for TP53 mutations to assess their predictive role in BEV response.
1 INTRODUCTION
Ovarian carcinoma (OC) represents the fifth leading cause of cancer-related death among women worldwide,1 due to its frequent presentation at an advanced stage at diagnosis and the development of chemoresistant recurrence, despite a high initial response to standard treatment consisting of cytoreductive surgery and platinum-based chemotherapy.2 Angiogenesis, one of the OC hallmarks, is implicated in cancer development and invasion, as well as in resistance to chemotherapy.3 Angiogenesis represents an appealing target for OC treatment, and, currently, the anti-vascular endothelial growth factor (VEGF) monoclonal antibody bevacizumab (BEV) is approved as first-line therapy, combined with platinum-taxane and as maintenance therapy for patients with advanced-stage OC.4, 5 In fact, the addition of BEV has significantly increased progression-free survival (PFS), particularly in high-risk OC patients, against moderate and manageable side effects.6 To date, there are no validated biomarkers predictive of BEV response in clinical use and patients are still selected for anti-angiogenic treatment only based on their clinical characteristics, such as stage, debulking status, and presence of ascites. However, it is crucial to identify new biomarkers to help in selecting patients who will benefit the most from BEV. The multicenter, Phase IV, single-arm MITO16A/MaNGO OV-2 trial was designed to fulfill this unmet clinical need, specifically to explore novel prognostic biomarkers for OC patients treated with carboplatin, paclitaxel, and BEV, followed by BEV maintenance, in first-line setting.7 The tumor-suppressor TP53 is the most frequently mutated gene in human malignant tumors,8 including OC.9 In addition to its well-known tumor-suppressor functions on cell cycle regulation, control of DNA repair mechanisms and activation of apoptotic pathways,10 TP53 is deeply involved in angiogenesis regulation through the degradation of hypoxia-induced factor-α11 and transcriptional control of VEGF-A.12 In the present study, we have investigated a large cohort of OC patients enrolled in the MITO16A-MaNGO OV-2 trial with two major aims: (i) to describe TP53 mutations in terms of type, function, site, and explore their prognostic role; and (ii) to determine p53 protein expression by immunohistochemistry (IHC) and investigate its prognostic potential.
2 MATERIALS AND METHODS
2.1 Study population and clinical samples
Three hundred ninety-eight patients were enrolled in the MITO16A/MaNGO OV-2 study.7 Formalin-fixed paraffin-embedded (FFPE) chemo-naïve tissue specimens were collected at clinical collaborating centers and sent to the INT G. Pascale of Naples, which centralized them. Sample quality check, tissue microarray (TMA) building, and nucleic acid extraction were performed by the INT G. Pascale of Naples, following recently published procedures.13
The flowchart reporting the causes of sample exclusion and the final number of tissues analyzed for p53 IHC and TP53 mutations is provided in Figure S1.
2.2 Determination and classification of TP53 mutations
Up to 200 ng of DNA obtained from FFPE specimens was used to construct next-generation sequencing (NGS) libraries targeting the TP53 gene. To this purpose, an amplicon-based mutation detection panel was designed for the TP53 gene (QIAGEN, USA), covering all coding and UTR exons, padding the exon-intron junctions by 10 bp to also detect splicing mutations. Briefly, up to 200 ng of tumor DNA was fragmented, barcoded, and enriched with the mentioned custom panel for the TP53 gene following the manufacturer's instructions (QIAseq targeted DNA, QIAGEN, USA). Libraries obtained were then amplified and checked for quality (4200 Tape Station System, Agilent Technologies, USA) and amount (Qubit dsDNA HS assay kit, Thermo Fisher, USA). Finally, the barcoded libraries were pooled and sequenced on an Illumina NextSeq 500 (Illumina, USA) sequencer, to reach an approximate coverage of 2500× for each sample. DNA extracted from blood samples was used as a control and subjected to the same preparation. Following sequencing, raw data from the instruments were aligned to the human reference genome (hg19) with “bwa”14 and de-duplication to remove artifacts using Universal Molecular Identifiers was performed with the “fgbio” software suite (https://github.com/fulcrumgenomics/fgbio). Seventy blood samples with high-quality data and no known germline variants were then pooled to create a panel of normal (PoN) with MuTect2. Variant calling of somatic TP53 variants was performed by running MuTect2 and VarDict15, 16 for each tumor sample against the previously prepared PoN. Variant effect and severity were calculated with the Variant Effect Predictor.17 Variants were then filtered to remove low-quality calls (depth <200×, variant allele fraction <10%) and known benign variants (general population frequency >1%, known benign variants in ClinVar). A second filtering step was performed to only include high-quality mutations with potential or known deleterious effects: known deleterious TP53 mutations were annotated with the IARC TP53 database (version 19, https://tp53.isb-cgc.org/) and additional missense mutations were screened for tumor driver potential with the Cancer Genome Interpreter (https://www.cancergenomeinterpreter.org/2018),18 only keeping mutations with a predicted driver value of “Tier 1” (high confidence) or “Tier 2” (medium confidence). The sequencing coverage and quality statistics for each sample are summarized in Table S1.
TP53 mutations were classified into three categories based on the type of mutation: (1) missense mutations, when a single nucleotide change codes for a different amino acid; (2) frameshift insertion or deletion or nonsense mutations resulting in a truncated, unstable protein; and (3) undetermined mutations, including splice site or intronic mutations or in-frame insertions and deletions. Moreover, TP53 mutations were divided into groups with respect to function: (1) missense mutations that result in “gain of function” that, as a consequence, not only abolish the tumor-suppressor activity but also endow the protein with oncogenic activities (oncomorphic mutations); (2) loss of function, frameshift insertion or deletion, or nonsense mutations resulting in a complete loss of tumor-suppressing activity; and (3) unclassified missense mutations that refer to single base substitutions not fulfilling oncomorphic criteria and that currently have an unknown function. “Gain of function” missense mutations (P151S, Y163C, R175H, L194R, Y220C, R248Q, R248W, R273C, R273H, R273L, and R282W) were defined according to Brachova et al.19, 20 and confer to the mutated protein an oncogenic phenotype, as demonstrated in in vitro and in vivo preclinical models.
2.3 TMA building, p53 IHC, and scoring
MITO16A/MaNGO OV-2 TMAs were created by selecting representative areas for each case from hematoxylin and eosin stained sections. The seven TMAs were constructed using an automated TMA (Galileo CK3500 TMA, ISENET, Milan, Italy) and contain a total of 358 tissue cores from 236 primary and 122 metastatic OC samples. Three 1-mm cores have been collected from different areas of each tumor block in order to overcome sample heterogeneity and the possible loss of tissue due to cutting. About 4-μm-thick sections were cut from TMAs and stained with anti-p53 (clone DO-7, 1:2, Agilent) antibody on a Bond Max automatic immunostainer (Leica Biosystems). Immunoreaction was revealed by using Novolink Polymer (Leica Microsystems), followed by diaminobenzidine as a chromogen and hematoxylin as a nuclear counterstain. Stained slides were acquired using an Aperio C2 digital scanner and ScanScope software, and images were taken as a snapshot by using ImageScope, RRID:SCR_014311. Nuclear immunoreactivity was evaluated by two independent investigators (MB and LA), blinded for clinicopathological characteristics and p53 gene mutational data, and an established scoring system was applied.21 In cases of discrepant scoring results, a consensus meeting with a third investigator (EK) was performed to solve the discordance. In detail, an abnormal p53 pattern includes (i) tumors with “overexpression” (OE), which was attributed to cases with 60%–100% of tumor cells showing intense nuclear p53 staining, or (ii) tumors completely negative for p53 in the presence of positive internal controls, defined as having a pattern of “complete absence,” so-called “null” (CA). Strong and diffuse (OE) p53 immunoexpression plus the CA of staining are interpreted as aberrant/abnormal p53 expression, as established previously.21 The normal or wild-type (WT) pattern was characterized by heterogeneous p53 staining, an alternating patchy pattern with negative staining, more commonly weak and focal.
2.4 Statistical analysis
Quantitative variables were described as median and interquartile range, while qualitative variables were reported as absolute numbers and relative frequency. The associations between biomarker (TP53 mutation and type/function classification) and the clinical prognostic factors were investigated using a chi-squared test. The prognostic effect of TP53 (mutation, type, and function) was evaluated using PFS and overall survival (OS) as endpoints. PFS was defined as the time elapsing from inclusion in the study to the first occurrence of either death for any cause or disease progression. OS was defined as the time elapsing between inclusion in the study and death for any cause. Kaplan–Meier curves were drawn for PFS and OS, the medians of survival were calculated and comparison was performed by a two-sided log-rank test. Furthermore, the prognostic role of TP53 on both PFS and OS was investigated using univariable and multivariable Cox proportional hazard models. Proportionality assumption was assessed using the Schoenfeld residuals. For each model, hazard ratios (HRs) and 95% confidence intervals (CIs) were reported. A multivariable model was performed using as covariates: Eastern Cooperative Oncology Group performance status (PS) (0 vs. 1–2), residual disease (none; ≤1 cm; >1 cm + not operated), The International Federation of Gynecology and Obstetrics (FIGO) stage (III vs. IV), and tumor histology (high-grade serous vs. other). Covariates were chosen according to the model defined in the manuscript reporting the clinical results of the trial.7 Data were analyzed using R software version 4.1.1 (R Foundation for Statistical Computing, Vienna, Austria).
3 RESULTS
3.1 Patient cohort
Among 398 patients enrolled in the MITO-16A/MaNGO-OV2 study, the following patients were excluded from the final p53 IHC evaluation: 13 patients because the samples were not available for research; 12 patients who underwent neo-adjuvant chemotherapy and could not provide treatment-naïve tissue; 15 because of inadequate tumor sample; 42 because the samples had inadequate morphology for TMA; 5 because tumor samples were exhausted after nucleic acid extraction. Samples from 311 patients were used for TMA construction and underwent p53 immunohistochemical staining (Figure S1).
For NGS, 25 out of 311 patients were excluded because the tumor sample was inadequate for DNA extraction. Of 286 remaining patients, the TP53 mutational status was investigated in 202 patients by means of NGS (Figure S1).
3.2 TP53 mutational analysis
For 197 out of 202 patients, TP53 mutational status was successfully determined. This cohort did not show any significant difference in terms of clinicopathological characteristics from the whole trial population (n = 398) (Table S2). Among 197 patients, 180 (91.3%) had tumors harboring TP53 mutation, while the remaining 17 (8.7%) had TP53 WT neoplasms. Table 1 describes the different types of TP53 mutations detected, with missense mutations representing the most common (n = 119, 66.1%), followed by truncating mutations (n = 51, 28.3%), mostly frameshift insertions and deletions (n = 34, 18.8%), and nonsense mutations (n = 16, 8.9%). Finally, there were 10 undetermined mutations, consisting of in-frame insertions and deletions, splice site, and intronic mutations. A total of 146 out of 180 (81.1%) hotspot mutations were observed, with the most common occurring at codons R273 (15 out of 146, 10.3%), R248 (11 out of 146, 7.5%), Y220 (8 out of 146, 5.5%), and R175 (6 out of 146, 4.1%). A detailed description of all detected TP53 mutations (in terms of variant types, function, domain, and allele frequency) with information on hotspot mutations is reported in Table S3.
| Type of mutation | ||||
|---|---|---|---|---|
| Characteristic | Missense (n = 119) | Truncating (n = 51) | Undetermined (n = 10) | Total cohort (n = 180) |
| Type of function, n (%) | ||||
| Gain of function | 43 (36.1) | 0 (0) | 0 (0) | 43 (23.9) |
| Unclassified | 76 (63.9) | 0 (0) | 4 (40.0) | 80 (44.4) |
| Loss of function | 0 (0) | 51 (100) | 6 (60.0) | 57 (31.7) |
| DNA domain, n (%) | ||||
| At | 1 (0.8) | 9 (17.6) | 0 (0) | 10 (5.6) |
| Ct | 1 (0.8) | 7 (13.7) | 0 (0) | 8 (4.4) |
| DNAb | 117 (98.3) | 35 (68.6) | 6 (60.0) | 158 (87.8) |
| Splice/Intron | 0 (0) | 0 (0) | 4 (40.0) | 4 (2.2) |
| Variant classification, n (%) | ||||
| Missense_Mutation | 119 (100) | 0 (0) | 0 (0) | 119 (66.1) |
| Frame_Shift_Del | 0 (0) | 25 (49.0) | 1 (10.0) | 26 (14.4) |
| Frame_Shift_Ins | 0 (0) | 8 (15.7) | 0 (0) | 8 (4.5) |
| In_Frame_Del | 0 (0) | 1 (2.0) | 4 (40.0) | 5 (2.8) |
| In_Frame_Ins | 0 (0) | 1 (2.0) | 1 (10.0) | 2 (1.1) |
| Nonsense_Mutation | 0 (0) | 16 (31.4) | 0 (0) | 16 (8.9) |
| Intronic_Mutation | 0 (0) | 0 (0) | 1 (10.0) | 1 (0.6) |
| Splice_Site | 0 (0) | 0 (0) | 3 (30.0) | 3 (1.6) |
In Table 1, the different functions of TP53 mutations are displayed and classified as: (i) gain of function (n = 43, 23.9%), that either abolish p53 WT functions or confer novel oncogenic properties and are represented by most, but not all, missense mutations; (ii) loss of function (n = 57, 31.7%), that significantly abate p53 tumor suppressor activity and are represented by all nonsense and frameshift mutations; and (iii) unclassified (n = 80, 44.4%), mostly missense mutations with unknown or undefined function. Regarding mutation site, as expected, the majority of TP53 mutations (158 out of 180, 87.8%) were localized in the DNA binding domain, covering exons 4–8, while a minority of them were found in the amino- (5.6%) or carboxy-terminal domains (4.4%) or in the splice (1.6%) or intron regions (0.6%) (Table 1 and Figure S2).
3.3 Association of TP53 mutational status with clinical features and survival
TP53-mutated tumors were significantly associated with high-grade serous histology (p = .0025) and with no other clinicopathological characteristics (Table 2). Type of mutation (missense, truncating, and undetermined) and function (gain of function, loss of function, and unclassified) were also significantly correlated with tumor histology (Tables S4 and S5).
| TP53 mutational status | ||||
|---|---|---|---|---|
| Characteristic | Total cohort (n = 197) | Wildtype (n = 17) | Mutated (n = 180) | p |
| Age elderly, n (%) | .41 | |||
| <65 | 142 (72.1) | 14 (82.4) | 128 (71.1) | |
| ≥65 | 55 (27.9) | 3 (17.6) | 52 (28.9) | |
| FIGO Stage, n (%) | .49 | |||
| III | 165 (83.8) | 13 (76.5) | 152 (84.4) | |
| IV | 32 (16.2) | 4 (23.5) | 28 (15.6) | |
| ECOG, n (%) | .75 | |||
| 0 | 157 (79.7) | 13 (76.5) | 144 (80.0) | |
| 1/2 | 37 (20.3) | 4 (23.5) | 36 (20.0) | |
| Tumor histology, n (%) | .0025 | |||
| HG serous | 171 (86.8) | 10 (58.8) | 161 (89.4) | |
| Other | 26 (13.2) | 7 (41.2) | 19 (10.6) | |
| Residual, n (%) | .29 | |||
| None | 76 (38.6) | 4 (23.5) | 72 (40.0) | |
| ≤1 cm | 44 (22.3) | 6 (35.3) | 38 (21.1) | |
| >1 cm/not operated | 77 (39.1) | 7 (41.2) | 70 (38.9) | |
- Abbreviations: ECOG, Eastern Cooperative Oncology Group; FIGO, The International Federation of Gynecology and Obstetrics; HG, high-grade.
As a result of the univariable survival analysis, the presence of a TP53 mutation was significantly prognostic for a favorable outcome in our cohort of patients. In particular, a significant association with an increased OS was observed for TP53-mutated tumors compared to WT cases (HR = 0.48, 95% CI = 0.24–0.97, p = .039), as represented in Figure 1A (median OS, 41.2 vs. 30.6 months, respectively). The significantly positive prognostic value of mutated TP53 on OS was also maintained when a multivariable Cox survival analysis was applied, adjusted for PS, FIGO stage, residual tumor and histology (HR = 0.46, 95% CI = 0.22–0.96, p = .039, Figure 2A). Differently, no significant difference in PFS was observed between patients with TP53 mutated tumors compared to WT by univariable analysis (p = .21, Figure S3).
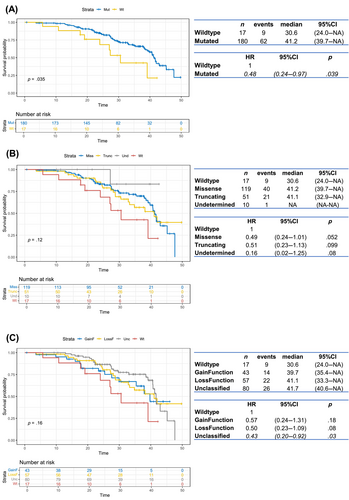
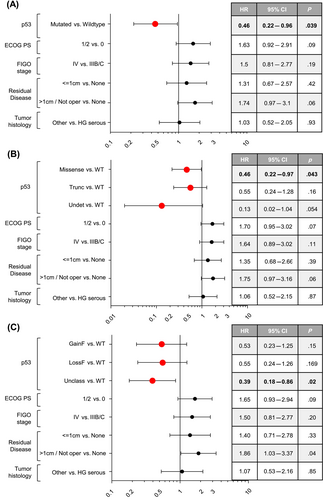
Then, the univariable survival analysis was performed considering the four groups: (1) WT, (2) missense mutations, (3) truncating mutations, and (4) undetermined mutations. We observed that the presence of a missense mutation was of borderline significance for improved OS (HR = 0.49, 95% CI = 0.24–1.01, p = .052, Figure 1B). This trend becomes statistically significant (HR = 0.46, 95% CI = 0.22–0.97, p = .043) in multivariable survival analysis, where missense mutations were associated with a favorable OS compared to WT (Figure 2B). Finally, we analyzed survival in patients harboring tumors with TP53 mutations classified according to function: (1) WT, (2) gain of function, (3) loss of function, and (4) unclassified. The presence of an unclassified mutation was indicative of an improved OS both in univariable (HR = 0.43, 95% CI = 0.20–0.92, p = .03, Figure 1C) and multivariable analysis (HR = 0.39, 95% CI = 0.18–0.86, p = .02, Figure 2C).
3.4 p53 immunohistochemical results
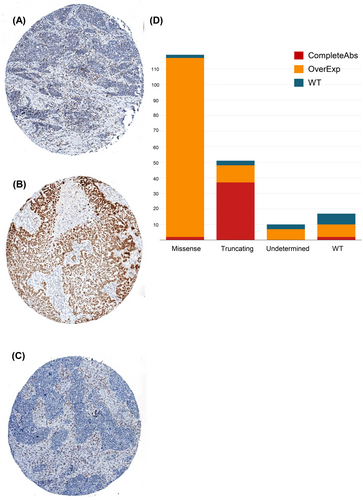
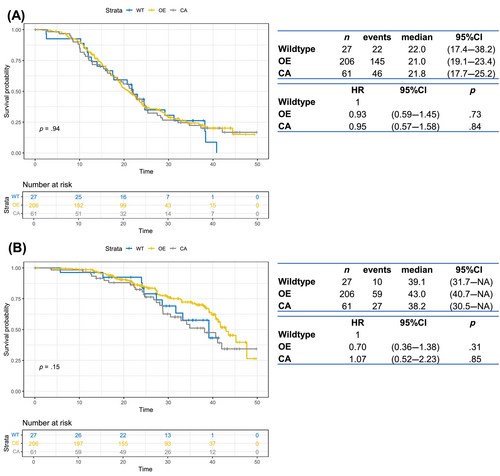
Out of 311 patients stained for p53 protein expression, 17 were excluded due to technical problems. Therefore, 294 patients with p53 immunostaining were available for further evaluation and analysis (Figure S1). Patients in the p53 cohort (n = 294) and in the whole MITO-16A/MaNGO-OV2 trial population (n = 398) did not show any difference in terms of clinicopathological characteristics (Table S2). p53 abnormal immunostaining was observed in 267 out of 294 (90.8%) OCs, while the remaining 27 (9.2%) showed a WT pattern (Figure 3A). Specifically, 206 cases showed an OE pattern and 61 tumors displayed CA staining (Figure 3B,C).
3.5 Association of p53 immunohistochemical results with clinicopathological features and survival
Abnormal p53 immunohistochemical expression was significantly associated with high-grade serous histology, with both OE and CA staining enriched in this subset of patients (p < 0.0001, Table S6). Applying a univariable survival analysis, no significant difference was observed in PFS according to immunohistochemical staining (Figure 4A). We observed an HR of 0.70 for OS in patients with p53 IHC overexpression compared to p53 WT patients, which however did not reach statistical significance (p = .31, 95% CI = 0.36–1.38), as displayed in Figure 4B.
3.6 Association of TP53 mutation status with p53 IHC
For 197 patients, both TP53 mutational status and p53 immunohistochemical staining results were available. As reported in the literature, tumors with truncating mutations generally showed p53 CA staining (72.5%), cases harboring missense mutations were predominantly p53 OE (96.6%), while WT tumors showed focal, weak p53 staining, as shown in Figure 3D. Overall, the degree of concordance between protein expression and mutational analysis was 91% (Table S7). Discordant cases were a limited number, only 18 out of 197 (9%), mainly present in the WT group (Figure 3D). The detailed list of 18 discordant cases is represented in Table S8.
4 DISCUSSION
The treatment approach for newly diagnosed advanced OC, consisting of primary debulking surgery followed by carboplatin and paclitaxel chemotherapy, has remained substantially unchanged for more than 40 years.22, 23 Recently, the introduction of targeted therapies such as poly (ADP-ribose) polymerase inhibitors (PARP-i) and BEV has shown efficacy in delaying cancer progression and they are currently employed as effective maintenance strategies in advanced OC. Notably, unlike several other targeted therapies, no pre-treatment biomarkers of response to BEV are available in clinical practice so far, making their identification a pressing clinical need.
In the context of the MITO16A/MaNGO OV-2 trial, we investigated the tumor-suppressor TP53, since it is frequently mutated in OC, involved in angiogenesis and could potentially affect the prognosis of patients treated with BEV. Herein, we demonstrate that the presence of TP53 mutations is associated with increased OS in OC patients treated with chemotherapy plus BEV in the upfront setting. Of note, this association holds true in multivariable analysis and remains an independent predictor of favorable OS after adjusting for all other clinical variables. When we performed the survival analysis based on the type and function of TP53 mutations, we observed that patients harboring missense mutations, in particular unclassified variants of unknown function, were characterized by a significant improvement in OS. As previously reported, different TP53 mutations have distinct functions and, consequently, drive cancers with divergent outcomes.24 It can be noticed that in our analyses the biomarker is statistically significant in the multivariable, but not in the univariable model. This could be explained by taking into account possible interactions between variables (e.g., a biomarker might become significant when it interacts with other variables), the increased amount of explained variability that may make it easier to detect the effect of the biomarker, or the presence of confounding variables that when considered in the model enable the effect of the biomarker to be detected more clearly.25 In addition, the presence of statistical significance in PFS but not in OS suggests that the occurrence of TP53 mutations could positively affect patients' OS (considering future treatments and differences in observation times), although not necessarily disease progression. Furthermore, GCIG Fifth Ovarian Cancer Consensus Conference stated that OS is the preferred primary endpoint, considering also potential confounding factors such as crossover and long post-progression survival.26
Several studies have investigated the correlation between TP53 mutations and patient outcome or response to chemotherapy in OC, showing conflicting results.24, 27, 28 However, they have explored the prognostic and predictive role of TP53 mutations only in cohorts of OC patients treated with primary debulking surgery followed by standard carboplatin plus paclitaxel chemotherapy. To our knowledge, our study is the first to report a prognostic role for TP53 mutations in OC patients receiving BEV in addition to standard chemotherapy. Nevertheless, our results are in line with previous clinical studies,29, 30 showing a significant survival improvement in patients with TP53-mutated advanced cancer treated with a BEV-containing chemotherapy regimen. Similarly, Hsu et al.31 reported TP53 missense mutations associated with prolonged PFS in metastatic colorectal cancer treated with a BEV-based therapy. A recent paper32 reported that the presence of a TP53 mutation confers a survival advantage in endometrial cancer patients when BEV, but not temsirolimus, is added to chemotherapy in the upfront setting. Consistent with our results, the authors found that missense mutations, most notably unclassified variants, were predictive of favorable outcome only in BEV-treated patients.33 The biological mechanism underlying the observed association between patients harboring TP53-mutated tumors and longer survival when treated with BEV remains not fully elucidated. Beyond its commonly reported involvement in DNA repair, cell cycle arrest and induction of apoptosis, growing evidence suggests that p53 could also be implicated in the inhibition of angiogenesis, that is, repressing the transcription of VEGF-A and other proangiogenic mediators.12, 34 When the transcriptional control of WT p53 is inactivated by somatic mutations in cancer, VEGF-A mRNA expression is upregulated, resulting in more vascularized tumors that might be efficiently targeted by BEV treatment. The association between TP53 mutation and VEGF-A expression has been reported in different cancers, independently of their tissue of origin.35, 36 The present investigation supports the hypothesis that, similarly to what has been observed in other tumors, TP53 mutation could represent a biomarker predicting a survival benefit in patients with OC treated with BEV. Since optimized p53 IHC is considered a reliable surrogate of mutation analysis,37 we performed and scored p53 immunostaining on tissues of OC patients included in the MITO16A/MaNGO OV-2 trial, correlating it with sequencing data and prognosis. We found a degree of concordance of 91% between protein expression by IHC and mutational analysis, in line with the literature.37, 38 The highest concordance was present in tissues harboring missense mutations, which showed p53 IHC OE in 96.6% of cases. Despite the optimal concordance between the two methods, when we performed survival analysis, patients with p53 OE showed only a trend toward improved OS. This finding is not surprising since the survival advantage associated with BEV therapy seems driven by unclassified variants in the context of missense mutations and IHC could not discriminate among different missense mutations (gain of function and unclassified). Our results indicate that specific TP53 mutations could represent an independent prognostic factor in patients treated with chemotherapy plus BEV, supporting the assessment of TP53 mutations by NGS on OC tissues at the time of diagnosis and when considering chemotherapy plus BEV in the first-line setting. A limitation of our study is the low number of TP53 WT cases, enriched in non-high-grade serous histotypes, which reflects the normal distribution of OC histotypes.2 Indeed, even if statistically significant in univariable and multivariable survival analysis, our results derive from two groups unbalanced in number and certainly require further confirmatory studies, increasing the sample size especially for TP53 WT patients that in our cohort are poorly represented.
Another limitation may concern the sample size, as this is a secondary study of biomarker analysis embedded in a clinical trial with a different and defined outcome.
In conclusion, we report for the first time a link between TP53 mutation and a favorable outcome in OC patients receiving upfront BEV plus chemotherapy, using the cohort of MITO16A/MaNGO OV-2 trial, which aimed at confirming the indications coming from the BEV registration studies in a real-life population. In particular, patients harboring unclassified missense TP53 mutations, which represent the majority of variants in our population, are characterized by a significant survival advantage, independently of the other clinicopathological features. The present findings require further studies in larger cohorts to confirm the independent prognostic potential of TP53 mutations in BEV-treated OC patients. Moreover, we plan to assess TP53 mutations in tissue samples collected within the MITO16B-MaNGo-OV2 randomized phase III trial39 and to evaluate their potential role as a predictive marker of response to BEV, as already documented for endometrial cancer.32, 33
BEV remains a key option for maintenance treatment in first-line therapy of OC, even in the era of PARP-i. Indeed, the benefit of PARP-i is particularly evident in patients with homologous recombination deficient (HRD) tumors, in which the added value of BEVA to PARP-i alone is unknown,40 therefore the identification of predictive biomarkers selecting patients who will really benefit from adding BEVA represents an important clinical need. Similarly, patients with tumors negative for HRD can receive PARP-i or BEV as maintenance therapy,41 but at the moment we have no discriminating factors to understand which is the best treatment, so also in this case reliable predictive biomarkers of response are urgently needed to formulate the correct first-line therapeutic algorithm. Our findings advocate further investigations to dissect the role of distinct somatic TP53 mutations in predicting response to BEV.
AUTHOR CONTRIBUTIONS
Eliana Bignotti: Conceptualization; investigation; methodology; supervision; validation; writing – original draft; writing – review and editing. Vittorio Simeon: Formal analysis; methodology; software; writing – original draft; writing – review and editing. Laura Ardighieri: Investigation; methodology; visualization; writing – original draft; writing – review and editing. Elisabetta Kuhn: Methodology; writing – original draft; writing – review and editing. Sergio Marchini: Methodology; writing – review and editing. Daniela Califano: Investigation; methodology; project administration; supervision; writing – review and editing. Sabrina Chiara Cecere: Investigation; writing – review and editing. Mattia Bugatti: Investigation; software; writing – review and editing. Anna Spina: Investigation; writing – review and editing. Giosuè Scognamiglio: Investigation; methodology; writing – review and editing. Lara Paracchini: Investigation; writing – original draft; writing – review and editing. Daniela Russo: Investigation; writing – review and editing. Laura Arenare: Data curation; writing – review and editing. Germana Tognon: Investigation; supervision; writing – original draft; writing – review and editing. Domenica Lorusso: Supervision; writing – review and editing. Luca Beltrame: Formal analysis; software; writing – review and editing. Maurizio D'Incalci: Writing – review and editing. Enrico Sartori: Writing – review and editing. Andrea De Censi: Writing – review and editing. Franco Odicino: Writing – review and editing. Francesco Perrone: Data curation; formal analysis; software; writing – review and editing. Paolo Chiodini: Data curation; formal analysis; writing – review and editing. Sandro Pignata: Conceptualization; funding acquisition; methodology; project administration; resources; supervision; writing – review and editing.
ACKNOWLEDGMENTS
We would like to thank all the patients who gave their consent to donate tumor samples for the purpose of this research. We are grateful to Alessandra Bono Foundation for their support. The MITO 16A-MaNGO OV2A is a multicenter national, academic trial sponsored by NCI Naples. Roche Italy granted the study with partial funding and Bevacizumab provision. The study was partially supported by AIRC, IG18921, IG25932 to SP and Ministry of Health RF-2016-02363995 and CO-2018-12367051 to SP, and Ministry of Health Ricerca Corrente L/3 to SP.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
ETHICS STATEMENT
The study (www.clinicaltrials.gov number: NCT01706120) was approved by ethical committees at each participating center. All the patients signed an informed consent to donate their samples for translational research before study entry.
Open Research
DATA AVAILABILITY STATEMENT
The p53 data set (IHC) with some deidentified individual participant data (i.e., baseline patient characteristics, treatment data, safety data, and follow-up data) is available at https://zenodo.org/records/8138609. Further details and other data that support the findings of this study are available from the corresponding authors upon request.