Assessing computational predictions of the phenotypic effect of cystathionine-beta-synthase variants
Present Address: Dago F. Dimster-Denk, Pionyr Immunotherapeutics, San Francisco, CA; Gaurav Pandey, Department of Genetics and Genomic Sciences and Icahn Institute for Data Science and Genomic Technology, Icahn School of Medicine at Mount Sinai, New York, NY; Emidio Capriotti, BioFolD Group, Department of Pharmacy and Biotechnology (FaBiT), University of Bologna, Bologna, Italy; Emanuela Leonardi, Department for Woman and Child Health, University of Padua, Italy; David Masica, AbbVie Inc., Redwood City, CA; Sean D. Mooney, Department of Biomedical Informatics and Medical Education, University of Washington, Seattle, WA; Predrag Radivojac, Khoury College of Computer Sciences, Northeastern University, Boston, Massachusetts; Janita Thusberg, Invitae, San Francisco, CA, USA; Susanna Repo, ELIXIR, Wellcome Genome Campus, Hinxton, Cambridge, UK; Mauno Vihinen, Department of Experimental Medical Science, Lund University, Lund, Sweden; Susanna Repo, ELIXIR, Wellcome Genome Campus, Hinxton, Cambridge, UK
Abstract
Accurate prediction of the impact of genomic variation on phenotype is a major goal of computational biology and an important contributor to personalized medicine. Computational predictions can lead to a better understanding of the mechanisms underlying genetic diseases, including cancer, but their adoption requires thorough and unbiased assessment. Cystathionine-beta-synthase (CBS) is an enzyme that catalyzes the first step of the transsulfuration pathway, from homocysteine to cystathionine, and in which variations are associated with human hyperhomocysteinemia and homocystinuria. We have created a computational challenge under the CAGI framework to evaluate how well different methods can predict the phenotypic effect(s) of CBS single amino acid substitutions using a blinded experimental data set. CAGI participants were asked to predict yeast growth based on the identity of the mutations. The performance of the methods was evaluated using several metrics. The CBS challenge highlighted the difficulty of predicting the phenotype of an ex vivo system in a model organism when classification models were trained on human disease data. We also discuss the variations in difficulty of prediction for known benign and deleterious variants, as well as identify methodological and experimental constraints with lessons to be learned for future challenges.
1 INTRODUCTION
One of the central challenges in biology is to determine the impact of genomic variation on the phenotype(s) of an organism. As the amount of genomic data increases and accumulates at an exponential rate, comprehensive, and accurate prediction algorithms are needed when biological experiments are expensive or difficult to execute (Fernald, Capriotti, Daneshjou, Karczewski, & Altman, 2011). Missense mutations are the most studied class of protein-altering variants; however, even today the algorithms often disagree on the pathogenicity prediction of the variants (Ioannidis et al., 2016). To determine the optimal use of each algorithm in different tasks, a thorough and unbiased methodological assessment is required. The ultimate aim is to attain a better understanding of the complex genotype–phenotype relationship, and, most importantly, provide the basis for clinical application to improve human health (Rost, Radivojac, & Bromberg, 2016). Since 2010, the Critical Assessment of Genome Interpretation (CAGI) experiment has been seeking to address these needs by evaluating bioinformatics tools developed for phenotype prediction from genomic variation data (Hoskins et al., 2017).
Cystathionine-beta-synthase (CBS, MIM# 613381) is an extensively studied vitamin-dependent enzyme involved in cysteine biosynthesis via the transsulfuration pathway. The molecular architecture of human CBS comprises an N-terminal heme-binding domain (residues 1–70), followed by the catalytic domain (residues 71–381), and a C-terminal regulatory domain (residues 412–551; Majtan et al., 2018). The heme domain, which is found only in mammalian forms of CBS, lacks any significant structural elements and exhibits significant sequence diversity. Changes in the heme's coordination environment can be transmitted to the active site ~20 Å away, leading to inhibition of CBS activity (Weeks, Singh, Madzelan, Banerjee, & Spiro, 2009). The central domain belongs to the family of pyridoxal-5′-phosphate (PLP)-dependent enzymes, with the PLP cofactor bound via a Schiff base to K119 in the CBS active site. The C-terminal domain, also known as the Bateman module, contains two consecutive CBS-motifs (residues 412–471 and 477–551) that form distinct binding sites for S-adenosyl-methionine (AdoMet) and enable CBS homotetramerization. Two high-affinity and four low-affinity AdoMet binding sites have been identified per CBS tetramer, with distinct roles proposed in the regulation and activation (Pey, Majtan, Sanchez-Ruiz, & Kraus, 2013). Catalytic and regulatory domains are joined by a flexible linker (residues 382–411) that is sensitive to proteolysis. Targeted proteolysis of CBS results in a truncated, dimeric, and more active form of the enzyme, adding another layer of CBS regulation (Skovby, Kraus, & Rosenberg, 1984; Zou & Banerjee, 2003).
Homocystinuria due to CBS deficiency (MIM# 236200) is an autosomal recessive inborn error of sulfur amino acid metabolism, characterized by increased levels of homocysteine in the urine (Mudd, Levy, & Kraus, 2001), myopia, osteoporosis, or other skeletal abnormalities. The estimated worldwide prevalence of homocystinuria is approximately 1 in 100,000 (Moorthie, Cameron, Sagoo, Bonham, & Burton, 2014). More than 160 different disease-associated variants have been identified in the CBS gene (http://cbs.lf1.cuni.cz/index.php). The majority of these are substitutions that do not involve catalytic residues, suggesting that their effect resides in structural or conformational perturbations leading to a misfolded protein (Majtan et al., 2018). About one-half of homocystinuric patients respond to high doses of pyridoxine, the soluble form of PLP (Mudd et al., 2001) and several mutations are clearly pyridoxine remediable (B6-responsive homocystinuria): p.A114V, p.R266K, p.R369H, p.K384E, p.L539S, and the most common substitution p.I278T, which accounts for ~20% of all homocystinuric alleles (Dimster-Denk, Tripp, Marini, Marqusee, & Rine, 2013; Moat et al., 2004; Skovby, Gaustadnes, & Mudd, 2010).
Since CBS is well studied and its ex-vivo mutation effects are easily quantified, it is a tractable system for investigating phenotype–genotype relationships, making it an attractive target for the CAGI challenges. Here we present an assessment of computational predictions on the effects of single amino acid substitutions in the function of CBS. In the CAGI1 (2010), CBS challenge, participants were asked to predict yeast growth rates when compared with wild-type yeast based on amino acid substitution information. This data set comprised 51 synthetic single amino acid substitutions within the human CBS coding region. In the CAGI2 (2010), CBS challenge, a larger set of variants (78 amino acid substitutions) that had been observed in patients with homocystinuria was provided for the predictors. In both challenges, participants were asked to submit predictions on the effect of the variants in the function of CBS at high (400 ng/ml) and low (2 ng/ml) cofactor (pyridoxine) concentration. Both CBS predictions were blinded experiments. At the time these challenges took place, the experimental effects of the mutations had not yet been published. Altogether 38 predictions from CAGI1 and CAGI2 were assessed. Methods employed varied from the purely structural to ones combining both structural and sequence conservation information or annotation, and from meta-predictors (models that use the predictions of other methods as features) to methods driven mainly by sequence, while one submission was a set of random predictions.
In general, deleterious mutations were better predicted than variants with minor or no effects on phenotype, with the hardest to predict effects often involving variants with weak sequence and structural signals. The use of distinct assessment criteria revealed differences in performance between methods, with methods integrating sequence, structure, and functional features performing best overall.
2 METHODS
2.1 Dataset
The CAGI1 CBS challenge included 51 synthetic single amino acid substitutions within the human CBS (Table 1), while the CAGI2 CBS challenge included 78 single amino acid variants that had been observed in patients with homocystinuria. The experimental data used in CAGI1 and CAGI2 were published after the challenges were closed by Dimster-Denk et al. (2013) and Mayfield et al. (2012), respectively. Only one variant, p.N228K, overlapped between CAGI1 and CAGI2. In addition, four positions (p.H65, p.L154, p.V354, and p.V371) overlap between the two challenges but the amino acid substitutions are different. The variant nomenclature refers to the human CBS cDNA (Refseq NM_000071.2).
| CAGI1 | CAGI2 | |||
|---|---|---|---|---|
| Nucleotide variant | Protein variant | Nucleotide variant | Protein variant | ClinVar (2019) |
| c.194A>T | p.H65L | c.194A>G | p.H65R | |
| c.250A>G | p.I84V | c.205G>C | p.A69P | |
| c.353T>G | p.V118G | c.209C>T | p.P70L | U |
| c.370C>G | p.L124V | c.233C>G | p.P78R | LP |
| c.370C>G, c.371T>C | p.L124A | c.253G>C | p.G85R | |
| c.379A>G | p.I127V | c.260C>A | p.T87N | |
| c.424A>G | p.I142V | c.262C>T | p.P88S | |
| c.424A>G,c.425T>C | p.I142A | c.302T>C | p.L101P | P/LP |
| c.425 T>A | p.I142N | c.304A>C | p.K102Q | |
| c.460C>G, c.461T>C | p.L154A | c.306G>C | p.K102N | LP |
| c.461T>C | p.L154P | c.325T>C | p.C109R | P/LP |
| c.529A>G | p.K177E | c.341C>T | p.A114V | P/LP |
| c.541C>G, c.542T>C | p.L181A | c.346G>A | p.G116R | LP |
| c.562A>G | p.I188V | c.361C>T | p.R121C | C |
| c.566T>C | p.V189A | c.362G>A | p.R121H | LP |
| c.629T>A | p.L210Q | c.362G>T | p.R121L | |
| c.640A>G | p.I214V | c.376A>G | p.M126V | |
| c.659T>G | p.L220R | c.384G>T | p.E128D | |
| c.684C>A | p.N228K | c.393G>C | p.E131D | U |
| c.718A>G | p.I240V | c.415G>A | p.G139R | P |
| c.718A>G, c.719T>C | p.I240A | c.429C>G | p.I143M | |
| c.721C>G | p.L241V | c.430G>A | p.E144K | P/LP |
| c.742C>G, c.743T>C | p.L248A | c.434C>T | p.P145L | P |
| c.755T>C | p.V252A | c.442G>C | p.G148R | LP |
| c.772G>C | p.G258R | c.451G>A | p.G151R | |
| c.800A>T | p.K267M | c.457G>C | p.G153R | U |
| c.799A>G | p.K267E | c.461T>A | p.L154Q | |
| c.811A>G | p.K271E | c.463G>A | p.A155T | |
| c.829A>C, c.830T>C | p.I277P | c.473C>T | p.A158V | |
| c.839T>C | p.V280A | c.494G>A | p.C165Y | P |
| c.856A>G, c.857T>C | p.I286A | c.502G>A | p.V168M | C |
| c.877C>G | p.L293V | c.539T>C | p.V180A | U |
| c.931A>G | p.I311V | c.572C>T | p.T191M | P |
| c.1012C>G, c.1013T>C | p.L338A | c.593A>T | p.D198V | |
| c.1023A>T | p.Q341H | c.671G>A | p.R224H | |
| c.1034T>C | p.L345P | c.676G>A | p.A226T | LP |
| c.1061T>G | p.V354G | c.683A>G | p.N228S | U |
| c.1067T>C | p.V356A | c.684C>A | p.N228K | |
| c.1073T>C | p.V358A | c.700G>A | p.D234N | P |
| c.1112T>C | p.V371A | c.715G>A | p.E239K | |
| c.1115T>C | p.V372A | c.770C>T | p.T257M | P/LP |
| c.1120C>G, c.1121T>C | p.L374A | c.775G>A | p.G259S | U |
| c.1147A>T | p.T383S | c.785C>T | p.T262M | P/LP |
| c.1153T>C, c.1154T>A, c.1155C>A | p.F385Q | c.796A>G | p.R266G | |
| c.1153T>C | p.F385L | c.797G>A | p.R266K | P/LP |
| c.1223G>T | p.W408L | c.824G>A | p.C275Y | |
| c.1268T>C | p.L423P | c.833T>C | p.I278T | P |
| c.1298A>T | p.H433L | c.862G>A | p.A288T | U |
| c.1370G>A | p.G457E | c.862G>C | p.A288P | |
| c.1468A>C | p.I490L | c.904G>A | p.E302K | LP |
| c.1646A>G | p.D549G | c.919G>A | p.G307S | P |
| c.959T>C | p.V320A | LP | ||
| c.992C>A | p.A331E | LP | ||
| c.992C>T | p.A331V | |||
| c.1007G>A | p.R336H | LP | ||
| c.1039G>A | p.G347S | LP | ||
| c.1046G>A | p.S349N | |||
| c.1055G>A | p.S352N | |||
| c.1058C>T | p.T353M | P/LP | ||
| c.1060G>A | p.V354M | |||
| c.1063G>C | p.A355P | |||
| c.1081G>A | p.A361T | |||
| c.1105C>T | p.R369C | U | ||
| c.1106G>A | p.R369H | U | ||
| c.1106G>C | p.R369P | |||
| c.1109G>A | p.C370Y | LP | ||
| c.1111G>A | p.V371M | LP | ||
| c.1126G>A | p.D376N | |||
| c.1150A>G | p.K384E | P | ||
| c.1173G>A | p.M391I | |||
| c.1265C>T | p.P422L | U | ||
| c.1301C>A | p.T434N | U | ||
| c.1304T>C | p.I435T | U | ||
| c.1316G>A | p.R439Q | C | ||
| c.1330G>A | p.D444N | P/LP | ||
| c.1367T>C | p.L456P | |||
| c.1397C>T | p.S466L | U | ||
| c.1572C>A | p.Q526K | |||
- Note: Positions are based on Refseq NM_000071.2. C denotes conflicting interpretations of pathogenicity, U uncertain significance.
The functionality of the variants was tested in an in vivo yeast complementation assay, where the human CBS allele is expressed in yeast and functionally complements the yeast ortholog, CYS4, which was removed from the chromosome. In this assay, growth is dependent upon the level of mutant human CBS function, and growth rates are expressed as a percentage relative to wild-type grown with the same amount of exogenous pyridoxine supplementation. An experimental standard deviation is also available based on 3–4 repeated assays. The assay was performed in the presence of high (400 ng/ml) and low (2 ng/ml) pyridoxine concentrations. For a detailed description of this assay, see Mayfield et al. (2012) and Dimster-Denk et al. (2013). Participants were asked to submit predictions on the effect of the variants on the function of CBS both in high and low-pyridoxine concentrations. The submitted prediction was requested as the percent of growth when compared with wild-type yeast, with a standard deviation. The predictions were then assessed against the percent of growth values actually measured for each substitution in the yeast assay.
2.2 Prediction assessment
The correlation between the predicted effects of the mutations and the actual effects serves as an initially simple but powerful measure to assess the accuracy of the prediction methods. Because the mutation data are not derived from a normal distribution, nonparametric tests were used to assess the methods. Both Spearman's rank correlation and Kendall's Tau correlation (KCC) were used to assess each algorithm's predictions against the observed growth rates. The root-mean-square deviation (RMSD) was also calculated to estimate the difference between experimental and predicted values. To assess the accuracy of the algorithms in a clinical sense, evaluation was also conducted against a binary version of the experimental growth rates. A threshold of 0 was chosen for the binary version and the performance was evaluated in terms of area under the ROC curve (AUC), sensitivity, specificity, accuracy, and positive/negative predictive value (Lever, Krzywinski, & Altman, 2016). For experimental data, the total number of negatives (no growth substitutions) were defined as N =TN+FP and the total number of experimental positives (growth detected) as P =TP+FN. All the performance indices are shown in Table S1. An overall ranking of the methods was defined as a mean ranking of three different measures (KCC, AUC, and RMSD) shown in Table S2. These three measures were chosen since they assess complementary aspects of prediction performance.
2.3 Bootstrapping
To assess the robustness of our comparison of different prediction models, and to identify any possible performance bias generated by outliers, we performed bootstrap simulation on measurements. 10,000 random samples were generated using sampling with replacement of the experimental data. The resulting evaluation metrics (KUC, AUC, and RMSD) were plotted as bar plots with error bars representing one standard deviation below and above the mean value for each metric generated through iterations of bootstrap sampling of data-points.
To estimate the statistical significance of pairwise prediction comparisons, we applied bootstrap simulation (as described above) to the metrics of interest (KCC, AUC, and RMSD). We then performed an all-vs-all comparison recording the p-values, and corrected for multiple hypothesis errors using the Bonferroni method.
2.4 Characterization of easy and hard to predict variants
We examined the mutation effects that were easiest and hardest to predict to determine whether they shared any common features. We first identified these mutations by summing the binary predictions (0 for no growth, > 0 for growth) at low-pyridoxine conditions across all methods for each variant. The variants with the lowest and highest summed scores were individually examined in terms of sequence, solvent-accessibility, and location within the CBS structure.
To determine the likelihood of a variant being benign or deleterious through sequence analysis, scores for amino acid substitutions were taken from the BLOSUM90 substitution matrix (Henikoff & Henikoff, 1992). Scores of − 1, 0, or 1 were classified as moderate substitutions, that is, substitutions with a likelihood of arising by chance in terms of evolution and therefore of unknown effect on CBS function. Scores > 1 were classified as conservative substitutions with a projected benign effect on CBS, while scores < −1 were classified as nonconservative, indicating a potentially deleterious effect on CBS function. Solvent accessible surface area (SASA) was calculated for the human CBS monomer (PDB id 4COO) using GetArea (Fraczkiewicz & Braun, 1998) and when different, dimer SASA results were noted. Secondary structure assignments and analysis were according to PDB id 4COO and visual inspection of the structure.
2.5 Method uniqueness in prediction results
For CAGI2, evaluation of the specific contribution of each prediction to the variance with experimental results was addressed using a multiple linear regression model. First, a multiple linear regression model was built with the best methods from each group. The top method from each group was chosen based on the highest adjusted R2 values of every single method, to exclude predictions using modified versions of the same methods. The final methods included in the model were SID#16, 23, 26, 27, 29, 34, 36, and 41. Subsequently, methods were removed one at a time, and the linear regression equation was recalculated. The contribution of each method to the model was estimated from the delta adjusted R2 values. SID#25 was excluded from the model as it lacked predictions for 10 substitutions.
3 RESULTS
3.1 CAGI1 challenge
In the CAGI1 CBS challenge, participants were asked to submit predictions to assess the impact of 51 single amino acid substitutions upon the function of the human CBS enzyme in both high (400 ng/ml) and low (2 ng/ml) pyridoxine concentrations. The function of the variants had been experimentally tested in an in vivo yeast complementation assay (Dimster-Denk et al., 2013). Twenty predictions from 13 groups were submitted to this challenge (Table 2), which were assessed blindly. A summary of each method is described in Supporting Information. Of the 13 participating groups, nine submitted one prediction, two contributed two, one submitted three and one provided four different submissions. Some methods used sequence-only or structure-only information, some employed meta-predictors, and others combined sequence, structural and annotation data. SID#17 (submission identification number) submitted only raw data without predictions and was excluded from the assessment. Most participants (18/19) provided predictions for both high and low-pyridoxine concentration; however, seven predictions did not distinguish between the different cofactor levels. The majority of the predictions did not include standard deviations (13/19), and most of the methods that included estimates of reliability for each prediction appeared to be arbitrary (constant values like σ = 5, 10, and 15; n = 5/7).
| Submission ID | Group ID | Program name | Program features | Reference |
|---|---|---|---|---|
| CAGI1 | ||||
| SID#1 | Lichtarge lab | Evolutionary action working version | Sequence and structure | |
| SID#2 | Bromberg lab | SNAP | Sequence, structure, and annotation | Bromberg and Rost (2007) |
| SID#3 | Wei lab | SAPRED | Structure | Ye et al. (2007) |
| SID#4 | Switch lab | FoldX | Structure | Schymkowitz et al. (2005) |
| SID#5 | Vihinen lab | PON-P | Meta-predictor | Olatubosun, Valiaho, Harkonen, Thusberg, and Vihinen (2012) |
| SID#6 | Vihinen lab | PolyPhen2 | Sequence, structure | Adzhubei et al. (2010) |
| SID#7 | Vihinen lab | SNPanalyzer | Sequence | Yoo, Lee, Kim, Rha, and Kim (2008) |
| SID#8 | Vihinen lab | Panther | Sequence | Thomas et al. (2006) |
| SID#9 | Casadio lab | IMutant3 | Structure and thermal stability | Capriotti, Fariselli, Rossi, and Casadio (2008) |
| SID#10 | Casadio lab | IMutant4 | Structure and thermal stability | |
| SID#11 | Casadio lab | IMutant baseline | Structure and thermal stability | |
| SID#12 | Forman lab | SDM | Sequence and structure | |
| SID#13 | BioFolD Unit | IMutant3 | Sequence and structure | Capriotti et al. (2008) |
| SID#14 | Karchin lab | Sequence and structure | ||
| SID#16 | Mooney lab | Meta-predictor | ||
| SID#18 | Forman lab | SDM | Sequence and structure | |
| SID#19 | Tavtigian lab | AlignGVGD | Sequence | Tavtigian, Byrnes, Goldgar, and Thomas (2008) |
| SID#20 | Tavtigian lab | AlignGVGD | Meta-predictor | |
| CAGI2 | ||||
| SID#16 | Bromberg lab | SNAP | Sequence and structure | Bromberg and Rost (2007) |
| SID#19 | Tosatto lab | Structure | ||
| SID#20 | Tosatto lab | Structure | ||
| SID#21 | Tosatto lab | Structure | ||
| SID#22 | Tosatto lab | Structure | ||
| SID#23 | Tosatto lab | Structure | ||
| SID#24 | Tosatto lab | D100 roll | Random | |
| SID#25 | Switch lab | Structure | ||
| SID#26 | Lichtarge Lab | Evolutionary Action | Sequence and structure | Katsonis and Lichtarge (2014) |
| SID#27 | Vihinen lab | PON-P | Meta-predictor | Olatubosun et al. (2012) |
| SID#28 | Vihinen lab | PON-P | Meta-predictor | Olatubosun et al. (2012) |
| SID#29 | Shatsky lab | Meta-predictor | ||
| SID#30 | Shatsky lab | Meta-predictor | ||
| SID#32 | Shatsky lab | Meta-predictor | ||
| SID#33 | Mooney lab | Meta-predictor | ||
| SID#34 | Sunyayev lab | Sequence and structure | ||
| SID#35 | Moult lab | SNPs3D SVM | Sequence and structure | Yue and Moult (2006) |
| SID#36 | Moult lab | SNPs3D SVM | Sequence, structure, and annotation | |
| SID#41 | Mooney lab | Meta-predictor |
3.1.1 Assessment across different performance metrics
No single evaluation measure can capture a method's performance; thus, various measures were used to assess the phenotype prediction programs, including Kendall's Tau correlation coefficient (KCC), precision/recall, accuracy, and root-mean-square deviation (RMSD), inter alia (Table S1). For KCC, most of the prediction methods display statistically significant correlation with experimental data at both pyridoxine concentrations (Figure 1). Methods SID#2, SID#5, and SID#20 showed strong correlation at both high and low cofactor concentrations. SID#7 was the second best at high pyridoxine concentration (τ = 0.50, p = 4.87 x 10−6); however, it showed little correlation at low cofactor concentration (τ = 0.25, p = .034). For RMSD, SID#5, which is a meta-predictor, was the best and second best at low and high cofactor concentration, respectively. The highest accuracy of 82.4% was achieved by three methods: SID#1, 6, and 20 (Table S1). The first two combined sequence and structure information, while SID#20 is a meta-predictor, integrating several prediction methods. SID#2, 3, 9, and 11 achieved 100% sensitivity, whereas other methods had the highest specificity (94–100%, SID#7 and SID#14) at both cofactor concentrations. The most sensitive models were mostly structure-based. The majority of the methods had good results for PPV, where the values varied from 65% to 100%; however, for NPV, the values displayed a much wider range (0–90%), meaning that the methods are better at predicting benign than deleterious variants.
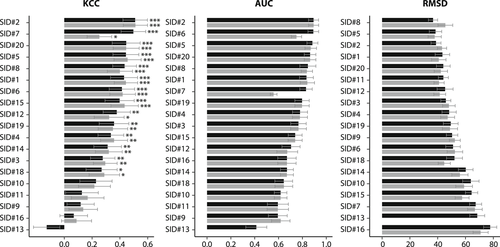
Kendall's Tau correlation coefficient (KCC), area under the ROC curve (AUC), and root-mean-square deviation (RMSD) for the phenotype prediction programs at high (black) and low (gray) cofactor concentration in the CAGI1 CBS challenge. Statistical significance of correlation scores is indicated with asterisks (*p ≤ .05, **p ≤ .01, ***p ≤ .001). Error bars represent one standard deviation below and above the mean value for each metric generated through iterations of bootstrap sampling of data-points
3.1.2 Overall ranking
To carry out an overall performance assessment, ranks of the prediction methods based on KCC, AUC, and RMSD were averaged to obtain the overall ranks of the methods (Table S2). This revealed SID#2 as the best performing method, having a mean rank of 2.2 across all measures, with SID#5 close behind (mean rank of 2.3). The first method is based on sequence information integrated with functional and structural annotations, while the other is a meta-predictor. One of the methods that did not perform as well, SID#16, was biased toward the prediction of low growth variants, whereas SID#13 was more conservative, with moderate to high growth predicted for most of the substitutions.
To estimate the robustness of the rankings, bootstrapping was performed (see Methods). Error bars for each metric were obtained by random resampling of the 51 variants 10,000 times. The rankings of the KCC, AUC, and RMSD for both high and low concentrations have only minor fluctuations, indicating that the prediction models are relatively robust (Figure 1). More than half of the predictions did better than the baseline method, SIFT (SID#15), which ranked 11th overall. Statistical significance estimation was performed for the metrics of interest (KCC, AUC, and RMSD) also using bootstrap simulation. None of the methods were significantly better than SIFT for KCC and AUC after Bonferroni correction (Figure S1 and S2). However for RMSD, SID#5, 11, and 20 outperformed SIFT at both cofactor concentrations even after Bonferroni correction (Figure S3).
3.1.3 Easy and hard to predict variants
We examined variants that were the easiest or hardest to predict based on the consensus output of all methods to determine whether they shared any common features (Figure 2b, Figure S7). At low cofactor concentration, there were overall 12% (2/17) deleterious variants and 18% (6/34) benign variants whose effects were predicted incorrectly by more than half of the methods, our definition of consensus.
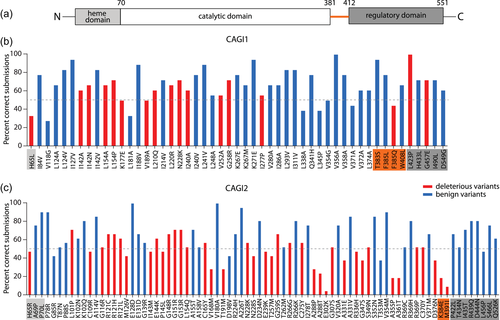
Consensus predictions for CBS. (a) CBS domain diagram, (b) CAGI1, (c) CAGI2. The percentage of correct predictions for deleterious (red) and benign (blue) variants is shown for each experimentally determined variant at low-pyridoxine concentration. Residues are shaded in the color of the corresponding domain, with the linker region highlighted in orange
The deleterious mutations that were easiest to predict at a low cofactor concentration were p.L154P, p.N228K, p.G258R, and p.G457E. The majority of these are nonconservative substitutions and are located within helices in the CBS structure (Table S3). The easiest to predict benign variants (low pyridoxine) were p.K271E, p.V356A, and p.T383S, with moderate conservation scores and were again mostly located within helices. Among the better predicted benign variants, the majority were partially or fully exposed to the solvent.
The hardest to predict deleterious variants at both high and low-pyridoxine concentrations were p.H65L and p.F385Q. p.H65 is located in the H1–H2 loop and axially coordinates the iron atom on one side of the heme plane, with C52 on the other, and mutation of either of these residues results in low-catalytic activity (Ojha, Wu, LoBrutto, & Banerjee, 2002). Interestingly, the functional impact of these variants was not easy to assess from sequence and structure comparisons. Although p.H65 and the sequence flanking it are locally conserved, the heme-binding domain itself (comprising approximately the first 70 N-terminal residues), with the exception of a short 5-residue helix, has no secondary structure elements and does not resemble other heme proteins in either primary sequence or tertiary structure (Kumar et al., 2018). p.F385Q is located in the H17–H18 loop that forms part of the linker connecting the N-terminal catalytic domain with the C-terminal regulatory domain, and lies within an aromatic cluster of residues p.Y381, p.F332, p.F334, p.F385, p.W390, and p.F396 enclosed by salt bridges p.R336-D388, p.K394-E302, and p.K384-E302 connecting helices H12–H14, H17, and H18. Erroneous coordination between aromatic residues can disrupt the extended pi-pi networks formed by aromatic clusters, thereby affecting protein stability and folding (Madhusudan Makwana & Mahalakshmi, 2015). In addition, both these variants involve nonconservative substitutions, and thus could be expected to have a deleterious effect on CBS function.
The hardest to predict benign variants (low pyridoxine) were surprising in that they involved nonconserved substitutions, so they could be expected to disrupt CBS function. In addition, within the structure, some were implicated in functionally relevant regions of CBS, such as the dimer interface (p.L345P) and the active site (p.V118G, adjacent to the PLP-ligating p.K119). All inaccurate consensus predictions of benign variants at low pyridoxine were for variants that confer sensitivity to reduced pyridoxine levels relative to the major allele (Dimster-Denk et al., 2013). The methods that correctly predicted all of these variants (SID#2, SID#3, and SID#9) displayed a broad spread both in features used (sequence, structure, and thermodynamics, respectively) and in overall performance (Table 2). In addition, SID#2 and SID#9 did not distinguish between high and low pyridoxine.
3.2 CAGI2 challenge
In the CAGI2 CBS challenge, 84 single amino acid variants that had been observed in patients with homocystinuria were collected and functionally tested in an in vivo yeast complementation assay (Mayfield et al., 2012). Seventy-eight had experimental values for both pyridoxine concentrations; 6 “hem1 rescue” variants were left out from the assessment due to absent/conflicting data (Table 1). Participants were again asked to submit predictions of the effect of the variants on the function of CBS both in high and low cofactor concentration. This challenge attracted 20 submissions from nine groups (Table 2) that were assessed without knowledge of the identity of the predictors. An overview of the methods is provided in Supporting Information. Four groups submitted one submission each, three groups submitted two each, and one group each contributed four and six predictions, respectively. Four groups participated in both CAGI1 and CAGI2 CBS challenges. As in CAGI1, features used to generate the predictions ranged from sequence- or structure-only information to meta-predictors and methods combining sequence, structural, and functional annotation data. SID#31 was excluded from the assessment due to its constant growth rate prediction of 100 for all substitutions. Almost all groups (17/19) provided distinct values for high/low cofactor concentrations. For this challenge, most submitters also provided standard deviations (13/19). Only one of the methods had arbitrary standard deviation values for all predictions (SID#26, σ = 10). In addition to prediction programs, reference results were obtained by submitting the mutations to the SNAP (SID#50) and SIFT (SID#51) public servers (Bromberg, Yachdav, & Rost, 2008; P. Kumar, Henikoff, & Ng, 2009).
3.2.1 Assessment across different performance metrics
The same assessment measures as in CAGI1 were used in CAGI2 (Methods). Looking at KCC, over half of the predictions were highly significant (Figure 3); however, even the best deviated substantially from experimental values. At both high and low-pyridoxine concentrations, methods SID#16 and SID#26 showed the strongest correlation with the experimental data and had also high AUC values (Figure 3). The latter was also the top predictor in terms of RMSD. In terms of accuracy, a structure-based method (SID#25) had the highest value (72%) at high and low-pyridoxine concentration (Table S2). At both cofactor concentrations, methods SID#16, 26, 34, and 41 had the highest sensitivity (100%). Most of these methods employed integrated sequence and structure information. SID#23 was the top method (high pyridoxine) with a 75% specificity, SID#27 and SID#28 scored 83% (low pyridoxine). SID#23 used structural data, whereas the other two are meta-predictors. In contrast to the CAGI1 CBS challenge, NPV showed higher median values than PPV (65 vs. 57%), implying that the probability of loss of function was slightly better predicted than the probability of having no or minor effects on the phenotype.
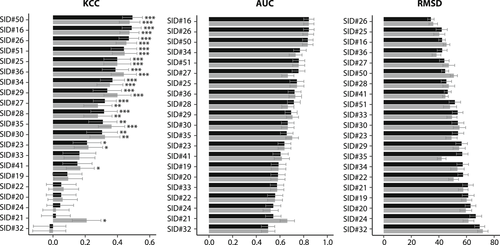
Kendall's Tau correlation coefficient (KCC), area under the ROC curve (AUC) and root-mean-square deviation (RMSD) for the phenotype predictions at high (black) and low (gray) cofactor concentration in the CAGI2 CBS challenge. Statistical significance of correlation scores is indicated with asterisks (*p ≤ .05, **p ≤ .01, ***p ≤ .001). Error bars represent one standard deviation below and above the mean value for each metric generated through iterations of bootstrap sampling of data-points
3.2.2 Overall ranking
As in the CAGI1 CBS challenge, we computed the overall ranks representing the performance of the submissions. Based on this criterion, the top methods in the CAGI2 CBS challenge were SID#26 and SID#16 with overall ranks of 1.8 and 2.3, respectively. Both methods utilized combined evolutionary information and structural features and ranked higher than the best baseline method (SID#50). For CAGI2, random resampling of the 78 variants 10,000 times generated error bars for each metric. As in CAGI1, the overall performance ranking remained unchanged. Methods that performed well generally resulted in smaller error bars, whereas methods that had only a few correct predictions exhibited a larger variance in performance as assessed by resampling (Figure 3).
Almost all of the methods performed better than the random predictor (SID#24), with the exception of SID#32 that ranked lower according to all assessment measures at both cofactor concentrations (Table S2). SID#32 is a random forest classifier that provided only binary outputs for both concentrations (no growth or growth, with values of 0 or 100, respectively). According to the bootstrap simulation, SID#16 and 26 performed significantly better than random at high pyridoxine concentration for all metrics (Figure S4–6). Two of the baseline methods, SNAP (SID#50) and SIFT (SID#51), had an overall ranking of third and sixth, respectively. None of the methods outperformed the baseline methods for KCC and AUC after Bonferroni correction (Figure S4, 5). For RMSD on the other hand, SID#26 did significantly better than SIFT at high cofactor concentration and SID#16 outperformed SNAP at low cofactor concentration.
3.2.3 Easy and hard to predict variants
The hardest and easiest to predict variants (Figure 2c, Figure S8) showed similar trends to those observed in the CAGI1 CBS challenge. As observed before, the inaccurately predicted benign variants (low pyridoxine) involved nonconservative substitutions and were located in loop regions of the CBS structure (Table S3). The most accurately predicted deleterious variants (low pyridoxine) involved a majority of nonconservative mutations of residues located within stable secondary structure elements (helices), while the hardest to predict deleterious predictions involved residues with moderate conservation scores mostly located within loops.
Within this last group, a number of mutants have been indirectly implicated in CBS function through involvement in homooligomerization, redox sensing, and regulation. p.A355 lies within helix H15, which is in turn sandwiched between H4 and strand beta3 at the CBS homodimerization interface. By introducing a kink in H15, the p.A355P mutant could potentially disrupt the folding in this region of the protein thereby impacting CBS function. Similarly, p.V168 is positioned at the homodimer interface while p.M391 is located within helix H18, a region of putative involvement in CBS homotetramer formation (Ereno-Orbea, Majtan, Oyenarte, Kraus, & Martinez-Cruz, 2013). p.A288 packs against p.W323 on strand beta6, and next to it, p.F324 packs against p.A360 in helix H15. p.A361 lies within interacting distance of p.C370, a residue that has been implicated in homocystinuria (Kraus et al., 1999) and proposed to modulate CBS function through interaction with an endogenous regulator such as nitric oxide (Eto & Kimura, 2002). The p.A361T mutant could therefore potentially interfere with a functionally relevant modification (e.g., S-nitrosylation) of p.C370. Similarly, modification of p.A288 could disrupt the pairing or orientation between beta5 and beta6, thereby potentially impacting the 272-CxxC-275 oxygen sensing motif of CBS, a redox active disulfide bond that allosterically controls CBS activity (Niu et al., 2018).
The most inaccurately predicted deleterious mutant, p.E302K, lies within interacting distance of one of the two active site loops (situated between helices H6 and H7). Recent studies have highlighted the importance of conformational flexibility of the loops defining the entrance to the catalytic site (Majtan et al., 2018).
3.2.4 Correlation between methods and unique contribution of different methods
To have a better understanding of the strengths and weaknesses of the different methods, we investigated the correlation between their predictions (Figure 4). Correlation heatmaps for high and low-pyridoxine concentration had negligible differences. The strongest predictor for method correlation appeared to be the relation to a single group (Table 2). For example, SID#27–28, SID#33&41, and all (SID#19–22) except one method (SID#23) were highly correlated among each other. However, SID#29–32 and 35–36 had higher correlations with other methods than among their own group. SID#29–32 were the only predictors that used simply two states (growth rates of 0 or 100). Interestingly, two of the best ranking methods (SID#16 and SID#26) were strongly correlated. In addition, SID# 27, 28, and 34 showed a strong correlation with the top prediction methods, although were based on different features.
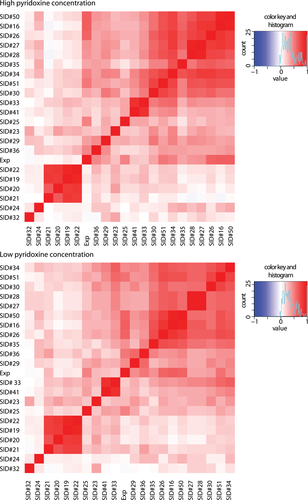
Spearman's rank correlation among methods and with experimental data (Exp) for high and low cofactor concentration. Each cell shows the correlation between two methods, with a color scale ranging from red (perfect correlation) to white (no correlation) and blue (perfect anti-correlation)
Baseline methods SNAP (SID#50) and SIFT (SID#51) showed strong correlation with the best performing predictions, which is partly expected as SID#16 was based on a version of the SNAP algorithm.
To assess the specific contribution of each method to the variance with experimental results in CAGI2, we applied a multiple linear regression model as described in Methods above. For high pyridoxine concentration, this revealed SID#16 and SID#36 as the most significant contributors (Δ adjusted R2 values of 0.053 and 0.041, respectively). At the same time, for low-pyridoxine concentration, SID#36 and SID#27 contributed the most (0.054 and 0.053, respectively). SID#36 is based on protein structure, sequence homology, and included functional information, whereas SID#16 combined evolutionary information with structural features, and SID#27 is a meta-predictor (Figure 5).
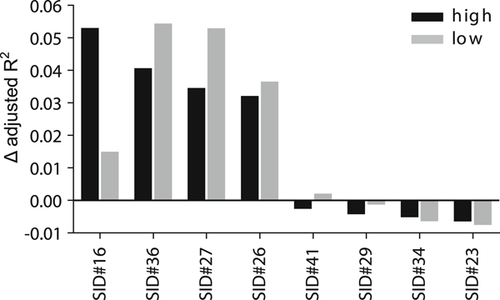
Δ adjusted R2 values of the methods from the linear regression model for high and low cofactor concentration, quantifying the contribution of each method to the proportion of total variance explained
4 DISCUSSION
4.1 Prediction features in relation to performance
In terms of prediction features, different methods performed well in distinct assessment measures. We observed that methods integrating sequence and structural information performed the best overall, ranking first or second (SID#2 in CAGI1 and SID#16 and SID#26 in CAGI2). Methods that used only structural information (SID#9 and 11 in CAGI1 and SID#19–20 in CAGI2) did not perform as well as those combining additional features. However, in terms of individual evaluation metrics, a structure-based method showed the highest accuracy of 72% (SID#25) in CAGI2. In CAGI1, SID#1 and 20 were the best at both cofactor concentrations, reaching accuracy of 82%. The first one combined structure and sequence data while SID#20 is a meta-predictor. In terms of sensitivity, most of the top-performing methods were structure-based in CAGI1 (SID#3, SID#9, and SID#11), while in CAGI2, the most sensitive algorithms combined structure with sequence data (SID#16, SID#26, and SID#34). At the same time, for specificity, almost all of the top methods were meta-predictors in CAGI2 (SID#27–28). These observations suggest that combining different features and methods would yield the best results, as has been indicated previously (Grimm et al., 2015; Tang & Thomas, 2016). Some methods are tailored to predict whether a variant affects the function of the protein in hand and others are optimized to determine whether a variant is pathogenic or benign in the clinical sense (Grimm et al., 2015; Katsonis et al., 2014; Pejaver, Mooney, & Radivojac, 2017).
The importance of integrating information from different sources is reflected in the most inaccurately predicted mutants that tended towards nonconserved substitutions, structural uncertainty, or both. The power of combining structural, sequence, and functional information was visible in CAGI2, where the overall performance of a structure and sequence combined method (SID#35) was improved significantly (by five ranks) with the inclusion of functional annotation data (SID#36). The latter was also the method that uniquely contributed the most predictive power of all methods at low-pyridoxine concentration. Another structure-based method (SID#25) that incorporated functional information (trained on the CAGI1 data set) also performed strongly. Methods trained on HGMD (Human Gene Mutation Database) mutations (SID#35–36) would be expected to perform well (Dong et al., 2015; Ioannidis et al., 2016; Pejaver et al., 2017). Interestingly, however, the best methods in CAGI2 CBS challenge showed variable training data, from no training to training on PMD (the Protein Mutant Database), HGMD, and CAGI1 CBS variant data (Supporting Information).
4.2 Limitations of the challenges
In terms of methodological limitations, most methods were developed to predict pathogenicity in humans or enzyme activity, not yeast growth or the effect of cofactor concentration on growth rate, something that could at least in part explain the difficulties they encountered in identifying the remediable class of variants (Supporting Information). So, while a meaningful distinction could be made in these challenges between growth and no growth under low-pyridoxine conditions, this was not the case for distinguishing between rescue (high pyridoxine) and no rescue (low pyridoxine) variants (Figure S9-12). Similarly, only qualitative comparisons could be made between CAGI1 and CAGI2, since the data sets differed in size and type, and only four groups participated in both challenges. Among these, one group used different versions of their method (SID#1 in CAGI1 and SID#26 in CAGI2), while two others did not make use of the CBS training data.
Another limitation of the assessment involved requesting standard deviation as estimates of reliability from predictors, as opposed to the more commonly employed confidence levels that most prediction methods provide. Consequently, some predictors did not provide these values, chose them arbitrarily, or provided large values with the result that they could not be reasonably used in these assessments.
These challenges also revealed a number of experimental limitations. Yeast CBS lacks the heme domain and is not regulated by AdoMet (Jhee, McPhie, & Miles, 2000), thereby engaging different pathways in the enzyme's regulation and physiological roles. In addition, overexpression can result in nonphysiological effects, including protein aggregation. These differences could help explain some of the inconsistencies observed in the experimental study in which yeast growth phenotypes did not match the clinical data (Mayfield et al., 2012). Although, only three positions from the heme domain were part of these two CBS challenges, with merely one position being problematic for the predictors (Figure 2). In a similar study, several variants identical to the ones used in these experiments resulted in contrasting yeast growth phenotypes (Wei, Wang, Wang, Kruger, & Dunbrack, 2010).
In addition, the clinical assessment of the majority of variants explored in this study has since changed. Of the 78 alleles described in CAGI2 as having been observed in patients with homocystinuria, only 30 are currently classified as pathogenic or likely pathogenic in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, accessed March 25, 2019), with an additional 16 annotated as being of uncertain significance or with conflicting interpretations of pathogenicity (Table 1). Eight substitutions (p.P78R, p.K102N, p.D234N, p.R266K, p.V320A, p.T353M, p.V371M, and p.D444N) out of 22 that showed experimental growth rates of ≥ 85% in CAGI2 are currently annotated as pathogenic or likely pathogenic. Most of the participants made accurate predictions for these “benign” variants (Figure 2). It is important to mention here that ClinVar had not yet been launched during the first two CAGI challenges. Also, not all the (likely) pathogenic CBS variants currently present in ClinVar have been collected from clinical testing, some are based on literature with no assertion criteria provided. Ideally, different functional assays should be applied, to increase the confidence in the observed phenotypic effect of the studied variant, because the function of a gene can differ in distinct organisms. Finally, mutations in cis with the ability to either suppress other pathogenic missense mutations or increase the severity of the clinical phenotype continue to be reported (de Franchis, Kraus, Kozich, Sebastio, & Kraus, 1999; Shan, Dunbrack, Christopher, & Kruger, 2001), raising the possibility that the incidence of double mutant alleles may be underestimated in homocystinuric patients.
5 CONCLUSION
CBS is a multifunctional enzyme with complex biology and intricate regulation that remains the object of much study. Our assessment of the CAGI1 and CAGI2 CBS challenges highlighted the strengths and weaknesses of different prediction features and approaches, as well as the need to address issues of methodological and experimental limitations. Both computational and experimental methods need to be tailored to the particular biological question under investigation to improve the predictive potential of the variant effect. It is hoped that future iterations of CAGI will see improvements on all these fronts.
ACKNOWLEDGMENTS
The authors are grateful to the CAGI organizers, data providers, participants, and would like to thank Pauline C. Ng for her contribution in the initial analysis of CAGI1. The CAGI experiment coordination is supported by NIH U41 HG007346 and the CAGI conference by NIH R13 HG006650. We are grateful to Robin Peters-Petrulewicz for a careful reading of the manuscript.
CONFLICT OF INTERESTS
The authors declare no conflicts of interest.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available to registered users from the CAGI web site https://genomeinterpretation.org/content/cagi-2011-results.