Deleterious de novo variants of X-linked ZC4H2 in females cause a variable phenotype with neurogenic arthrogryposis multiplex congenita
Abstract
Pathogenic variants in the X-linked gene ZC4H2, which encodes a zinc-finger protein, cause an infrequently described syndromic form of arthrogryposis multiplex congenita (AMC) with central and peripheral nervous system involvement. We present genetic and detailed phenotypic information on 23 newly identified families and simplex cases that include 19 affected females from 18 families and 14 affected males from nine families. Of note, the 15 females with deleterious de novo ZC4H2 variants presented with phenotypes ranging from mild to severe, and their clinical features overlapped with those seen in affected males. By contrast, of the nine carrier females with inherited ZC4H2 missense variants that were deleterious in affected male relatives, four were symptomatic. We also compared clinical phenotypes with previously published cases of both sexes and provide an overview on 48 males and 57 females from 42 families. The spectrum of ZC4H2 defects comprises novel and recurrent mostly inherited missense variants in affected males, and de novo splicing, frameshift, nonsense, and partial ZC4H2 deletions in affected females. Pathogenicity of two newly identified missense variants was further supported by studies in zebrafish. We propose ZC4H2 as a good candidate for early genetic testing of males and females with a clinical suspicion of fetal hypo-/akinesia and/or (neurogenic) AMC.
1 INTRODUCTION
Arthrogryposis multiplex congenita (AMC) has a prevalence of one case per 3,000–5,000 newborns. It is defined as multiple joint contractures that involve at least two different body areas before birth. AMC is a descriptive term and present in over 400 specific conditions (Hall, 1984, 2014). One known causal factor is decreased fetal movement in utero (fetal hypo-/akinesia; Hall, 2009). AMC can result from a single gene defect or a chromosomal abnormality and can be part of various syndromes. In these cases, autosomal dominant, autosomal recessive, and X-linked inheritance are possible (Hall, 2014). There are currently over 800 genes connected with either arthrogryposis or other types of early contractures with over 150 forms due to deleterious defects of X-linked genes (Hunter et al., 2015).
One form of X-linked arthrogryposis was first described in 1985 in six men from three generations of one family and referred to as Wieacker-Wolff syndrome (WRWF; MIM# 314580; Wieacker, Wolff, Wienker, & Sauer, 1985). All six affected had congenital contractures of the feet, slowly progressive predominantly distal muscle atrophy, visual dyspraxia, facial weakness, and intellectual disability (ID). In 2013, this family was reported to carry a pathogenic missense variant of the X-linked gene ZC4H2 (MIM# 300897) along with three additional unrelated families who carried ZC4H2 missense variants in males and females, an unrelated male with a de novo chromosomal inversion that truncated ZC4H2, and two unrelated females harboring heterozygous de novo ZC4H2 deletions (Hirata et al., 2013). In zebrafish, zc4h2 knockdown caused abnormal swimming and impaired alpha-motor neuron development, which could not be rescued by mutant proteins containing the pathogenic substitutions (Hirata et al., 2013).
In 2015, May et al. identified ZC4H2 variants in four additional families, including a large family with syndromic X-linked ID (XLID; MRXS4), previously described as having Miles-Carpenter syndrome (MCS; Miles & Carpenter, 1991) and characterized by XLID, exotropia, distal muscle wasting, microcephaly, congenital contractures, and low digital arches (May et al., 2015). Most recently, three simplex females with heterozygous de novo deleterious ZC4H2 were reported, including an early truncating nonsense variant and two microdeletions (Godfrey, Dowlatshahi, Martin, & Rothkopf, 2018; Okubo et al., 2018; Zanzottera et al., 2017). As a result of these findings, these various allelic syndromes are now referred to as ZC4H2-associated rare disorders (ZARD).
We here report 23 additional ZARD families and simplex cases with inherited or de novo pathogenic ZC4H2 variants plus three cases with publicly available information in DECIPHER (https://decipher.sanger.ac.uk/). We compared the genetic and clinical results with previously published families, thereby extending the molecular and clinical spectrum of ZARD throughout life, and discuss the broad clinical spectrum and its clinical variability in both males and females. Furthermore, we report a late adult-onset mild form of slowly progressive spastic paraplegia in ZC4H2 carrier females of one previously published family (Hennekam, Barth, Van Lookeren Campagne, De Visser, & Dingemans, 1991; Hirata et al., 2013). We emphasize that the ZARD phenotype in females with a pathogenic de novo ZC4H2 variant can be highly variable.
2 MATERIALS AND METHODS
2.1 Editorial policies and ethical considerations
The study was carried out in accordance with the Declaration of Helsinki and the protocol approved by the local ethical committees for clinical genetic investigations. Written informed consent was obtained for molecular genetic analysis, publication of clinical, radiological data, and photographs from all participants or their legal guardians.
2.2 Genetic studies
DNAs were extracted from peripheral blood, skin fibroblasts, buccal cells, or umbilical cord using standard procedures.
Families 1, 14, 16, and 18 (DECIPHER Patient IDs: 263304, 263305, 260529, 276496, and 296515) were recruited to the Deciphering Developmental Disorders (DDD) study. DNA samples were analyzed by the Wellcome Sanger Institute using array-CGH and whole exome sequencing (WES; Wright et al., 2015). For families 2, 8, 13, 15, 19, and 24 trio WES was performed as described in more detail in Supporting Information Materials and Methods, and for families 3 and 5 essentially as reported previously (Neveling et al., 2013). For families 4 and 6 all ZC4H2 coding exons were amplified by polymerase chain reaction (PCR) and Sanger sequenced with gene-specific primer pairs previously published (Hirata et al., 2013). Also for family 6, all TYR and OCA2 coding exons were amplified by PCR and Sanger sequenced. For family 7, see, Hennekam et al. (1991) and Hirata et al. (2013). For family 9 WES was performed for the index proband (Figure 1, V:1), his unaffected parents (III:1 and III:2) and two of his affected male relatives (III:5 and IV:2). For family 17 WES was performed for the index proband (Figure 1, II:1) in a research context. For families 3 (umbilical DNA of II:3), 10, 11, 12, 20, and 23 array-CGH was carried out, and for family 12 the breakpoints of the chromosomal deletion were fine-mapped by serial PCR and bidirectional Sanger sequencing (for further details see Supporting Information and Figure S1). For family 21, trio whole genome sequencing (WGS) was performed on peripheral blood DNA from the proband and both parents through the 100,000 Genomes Project (The 100,000 Genomes Project Protocol v3, Genomics England. doi:10.6084/m9.figshare.4530893.v3. 2017). For family 22, trio WGS was performed on saliva DNA at Baylor Genome Center as part of the NIH Gabriella Miller Kids First Research Program. For this family, the GeneMatcher and matchbox nodes of the Matchmaker exchange database were used to obtain collaborations using the search term ZC4H2 (Arachchi et al., 2018; Philippakis et al., 2015; Sobreira et al., 2017). For family 24 trio WES was performed at Fulgent Genetics, US.
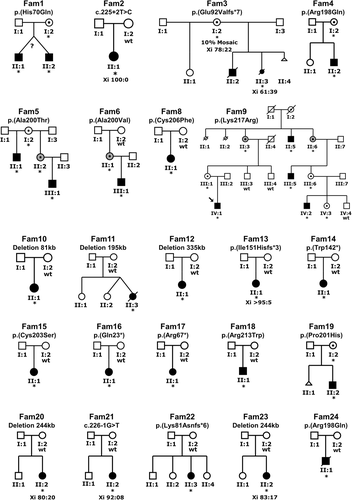
Pedigrees of the families with pathogenic ZC4H2 variants discovered by candidate gene approach, whole exome/genome sequencing and array-CGH. Families 1, 4–6, 8, 9, 15, 18, 19, and 24 with likely pathogenic ZC4H2 missense variants, families 2, 3, 13, 14, 16, 17, 21, and 22 with splicing, frameshift, and stop codon variants and families 10–12, 20, and 23 with a microdeletion removing the 5′ part of ZC4H2, Fam, family; *, variant present; wt, wild-type. For family 7 see Figure S2
The ZC4H2 variants identified by WES and WGS were subsequently confirmed in the patients using Sanger sequencing. Segregation analysis of the variants was performed using standard Sanger sequencing with gene-specific primers and for family 9 using restriction digestion of the specific PCR product (see Supporting Information for more details).
The novel ZC4H2 variants identified through this study have been submitted to the LOVD database (https://databases.lovd.nl/shared/genes/ZC4H2).
2.3 X-inactivation studies
For X-inactivation studies, DNA extracted from blood lymphocytes or skin fibroblasts was analyzed for the methylation sensitive site of FMR1 exon 1 (Carrel & Willard, 1996), the CAG-repeat of the Androgen Receptor (AR) gene (Allen, Zoghbi, Moseley, Rosenblatt, & Belmont, 1992), or of ZMYM3.
2.4 In silico analysis
The pathogenicity of missense variants was assessed using in silico tools including Combined Annotation Dependent Depletion (CADD, http://cadd.gs.washington.edu/) score (Kircher et al., 2014) and Provean (http://provean.jcvi.org/index.php; Choi & Chan, 2015).
2.5 Extraction of variants and clinical phenotypes from the literature and the DECIPHER database
We established a list of all published ZC4H2 variants reported in the literature as of January 2019 and three cases from DECIPHER, and included available clinical information in our analysis.
2.6 Knockdown and rescue experiments in zebrafish
Zebrafish were bred and maintained according to approved guidelines prescribed by the Committee on Use and Care of Animals at Aoyama Gakuin University (Japan). Zebrafish zc4h2 (GenBank NM_199642) cloned into pCR4-TOPO vector (Invitrogen) was used for cRNA synthesis as previously described (Hirata et al., 2013). Antisense morpholino oligonucleotides (MO) designed against the exon 2-intron 2 splice donor site (MO2) of zebrafish zc4h2 were used for knocking down zc4h2. Zebrafish embryos were injected with 5 ng of MOs at 1–2 cell stages and studied as published before (Hirata et al., 2013). For rescue experiments, the missense variants were introduced into the wild-type mouse Zc4h2 (RefSeq NM_001003916.2) pCS2+ expression construct by site-directed mutagenesis (primer sequences are provided in Table S1). Capped RNA was synthesized using mMESSAGE mMACHINE SP6 kit (Life Technologies) according to the manufacturer's protocol. Capped RNA (100 pg) was coinjected with MO2 (4 ng) into zebrafish embryos at 1–2 cell stages. At 2 days postfertilization, normal zebrafish embryos swim away rapidly (> 2 cm/s) upon tactile stimulation. The number of embryos that exhibited slow swimming (< 2 cm/s) following touch was counted.
2.7 ZC4H2 reference sequence
All ZC4H2 variants reported are based on GRCh37/hg19 and transcript isoform 1 (RefSeq NM_018684.3) with nucleotide (cDNA) numbering using +1 as the A of the ATG translation initiation codon in the reference sequence, with the initiation codon as codon 1.
3 RESULTS
3.1 Clinical features in the new patient cohort and in previously published families
We report on 23 novel families and simplex cases with likely deleterious ZC4H2 variants and on one previously published family, family 7 of this study (Hennekam et al., 1991; Hirata et al., 2013). The pedigrees of the novel families are shown in Figure 1 (Fam1–6 and Fam8–24) and of family 7 in Figure S2. Detailed clinical descriptions are presented in Supporting Information Results. In the clinical evaluation, we also included publicly accessible information of three females reported to DECIPHER and all families and simplex cases published to date (Godfrey et al., 2018; Hirata et al., 2013; Kondo et al., 2018; May et al., 2015; Okubo et al., 2018; Zanzottera et al., 2017). A compilation of the main (≥ 30%) clinical features of affected males and females from these 42 families is given in Table 1 and of the additional less common clinical features (< 30%) in Table S2. Percentages are related to the total number of probands who were clinically evaluated for a given feature (positive informative). In total, informative clinical data were collected for 87 out of 105 (83%) probands, including 44 out of 48 (92%) males and 43 out of 57 (75%) females using the Human Phenotype Ontology (https://hpo.jax.org/app/) scoring list. Their ages ranged from 23 weeks of gestation to > 70 years at the time of clinical genetic investigation. Families were recruited worldwide and were of Caucasian and Asian ethnicity.
| ZARD | Males total | Females | Females total | Males and females total | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ZC4H2 alterations | De novo and inherited | De novo | Inherited | De novo and inherited | De novo and inherited | ||||||||||||
| Category | Features | HPO | Total positive informative | Total informative | %* | Total positive informative | Total informative | %* | Total positive informative | Total informative | %* | Total positive informative | Total informative | %* | Total positive informative | Total informative | %* |
| Maximum informative | 44 | 48 | 92 | 23 | 25 | 92 | 20 | 32 | 63 | 43 | 57 | 75 | 87 | 105 | 83 | ||
| Growth | Short stature <3rd percentile | 4322 | 19 | 37 | 51 | 10 | 18 | 56 | 2 | 16 | 13 | 12 | 34 | 35 | 31 | 71 | 44 |
| Postnatal growth retardation | 8897 | 7 | 12 | 58 | 10 | 14 | 71 | 0 | 1 | 10 | 15 | 67 | 17 | 27 | 63 | ||
| Head and neck | Microcephaly <3rd percentile | 252 | 16 | 26 | 62 | 3 | 15 | 20 | 1 | 15 | 7 | 4 | 30 | 13 | 20 | 56 | 36 |
| Facial weakness-palsy | 10628 | 11 | 22 | 50 | 9 | 14 | 64 | 0 | 3 | 9 | 17 | 53 | 20 | 39 | 51 | ||
| High forehead | 348 | 6 | 11 | 55 | 7 | 12 | 58 | 1 | 2 | 50 | 8 | 14 | 57 | 14 | 25 | 56 | |
| Low-set ears | 369 | 12 | 19 | 63 | 8 | 15 | 53 | 1 | 9 | 11 | 9 | 24 | 38 | 21 | 43 | 49 | |
| Low-set, posteriorly rotated ears | 368 | 8 | 17 | 47 | 8 | 15 | 53 | 0 | 2 | 8 | 17 | 47 | 16 | 34 | 47 | ||
| Upslanting palpebral fissures | 582 | 10 | 21 | 48 | 1 | 12 | 8 | 0 | 1 | 1 | 13 | 8 | 11 | 34 | 32 | ||
| Ptosis | 1488 | 26 | 44 | 59 | 4 | 14 | 29 | 9 | 22 | 41 | 13 | 36 | 36 | 39 | 80 | 49 | |
| Deeply set eyes | 490 | 6 | 11 | 55 | 5 | 13 | 38 | 0 | 1 | 5 | 14 | 36 | 11 | 25 | 44 | ||
| Ocular motor apraxia | 657 | 8 | 12 | 67 | 4 | 10 | 40 | 1 | 2 | 50 | 5 | 12 | 42 | 13 | 24 | 54 | |
| Strabismus | 486 | 13 | 25 | 52 | 12 | 16 | 75 | 5 | 14 | 36 | 17 | 30 | 57 | 30 | 55 | 55 | |
| Anteverted nares | 463 | 13 | 22 | 59 | 9 | 13 | 69 | 3 | 9 | 33 | 12 | 22 | 55 | 25 | 44 | 57 | |
| Microretrognathia | 308 | 11 | 20 | 55 | 14 | 17 | 82 | 1 | 9 | 11 | 15 | 26 | 58 | 26 | 46 | 57 | |
| Long (flat) philtrum | 343 | 17 | 40 | 43 | 6 | 14 | 43 | 12 | 18 | 67 | 18 | 32 | 56 | 35 | 72 | 49 | |
| Broad alveolar ridges | 187 | 12 | 21 | 57 | 2 | 9 | 22 | 2 | 9 | 22 | 4 | 18 | 22 | 16 | 39 | 41 | |
| High-arched palate | 218 | 16 | 33 | 48 | 3 | 10 | 30 | 3 | 17 | 18 | 6 | 27 | 22 | 22 | 60 | 37 | |
| U-shaped upper lip vermilion - carpshaped mouth | 10806 | 20 | 42 | 48 | 5 | 13 | 38 | 0 | 11 | 5 | 24 | 21 | 25 | 66 | 38 | ||
| Cleft palate | 175 | 1 | 8 | 13 | 7 | 17 | 41 | 0 | 2 | 7 | 19 | 37 | 8 | 27 | 30 | ||
| Downturned corners of mouth | 2714 | 4 | 11 | 36 | 9 | 14 | 64 | 0 | 2 | 9 | 16 | 56 | 13 | 27 | 48 | ||
| Short neck (with limited rotation) | 470 | 12 | 37 | 32 | 12 | 14 | 86 | 1 | 19 | 5 | 13 | 33 | 39 | 25 | 70 | 36 | |
| Respiratory | Neonatal respiratory distress | 2643 | 16 | 34 | 47 | 5 | 16 | 31 | 0 | 13 | 5 | 29 | 17 | 21 | 63 | 33 | |
| Recurrent aspiration pneumonia | 2100 | 3 | 9 | 33 | 5 | 11 | 45 | 0 | 3 | 5 | 14 | 36 | 8 | 23 | 35 | ||
| Apnea | 2104 | 11 | 16 | 69 | 3 | 11 | 27 | 0 | 3 | 3 | 14 | 21 | 14 | 30 | 47 | ||
| Chest | Limited shoulder movement | 6467 | 7 | 16 | 44 | 12 | 14 | 86 | 1 | 2 | 50 | 13 | 16 | 81 | 20 | 32 | 63 |
| Narrow chest - narrow shoulders-thorax | 774 | 18 | 36 | 50 | 11 | 15 | 73 | 5 | 20 | 25 | 16 | 35 | 46 | 34 | 71 | 48 | |
| Abdomen | Feeding difficulties - poor feeding in infancy | 8872 | 19 | 30 | 63 | 12 | 17 | 71 | 2 | 13 | 15 | 14 | 30 | 47 | 33 | 60 | 55 |
| Encopresis - bowel incontinence | 40183 | 6 | 7 | 86 | 5 | 12 | 42 | 2 | 9 | 22 | 7 | 21 | 33 | 13 | 28 | 46 | |
| Genitourinary | Micropenis | 54 | 7 | 12 | 58 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | 7 | 12 | 58 |
| Cryptorchidism | 28 | 9 | 17 | 53 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | 9 | 17 | 53 | |
| Urinary incontinence | 20 | 2 | 4 | 50 | 9 | 12 | 75 | 5 | 12 | 42 | 14 | 24 | 58 | 16 | 28 | 57 | |
| Skeletal | Cervical kyphosis | 2947 | 20 | 40 | 50 | 7 | 18 | 39 | 5 | 19 | 26 | 12 | 37 | 32 | 32 | 77 | 42 |
| Thoracic kyphosis | 2942 | 19 | 40 | 48 | 7 | 18 | 39 | 5 | 18 | 28 | 12 | 36 | 33 | 31 | 76 | 41 | |
| Scoliosis | 2650 | 22 | 40 | 55 | 7 | 19 | 37 | 5 | 18 | 28 | 12 | 37 | 32 | 34 | 77 | 44 | |
| Hip contracture | 3273 | 15 | 32 | 47 | 15 | 19 | 79 | 2 | 21 | 10 | 17 | 40 | 43 | 32 | 72 | 44 | |
| Congenital hip dislocations - subluxation | 1374 | 14 | 32 | 44 | 11 | 19 | 58 | 2 | 21 | 10 | 13 | 40 | 33 | 27 | 72 | 38 | |
| Short limbs | 9826 | 7 | 17 | 41 | 3 | 14 | 21 | 6 | 10 | 60 | 9 | 24 | 38 | 16 | 41 | 39 | |
| Arthrogryposis multiplex congenita | 2804 | 31 | 45 | 69 | 23 | 25 | 92 | 8 | 18 | 44 | 31 | 43 | 72 | 62 | 88 | 70 | |
| Knee flexion contracture | 6380 | 26 | 38 | 68 | 13 | 19 | 68 | 10 | 21 | 48 | 23 | 40 | 58 | 49 | 78 | 63 | |
| Elbow flexion contractures | 2987 | 19 | 35 | 54 | 10 | 17 | 59 | 10 | 21 | 48 | 20 | 38 | 53 | 39 | 73 | 53 | |
| Wrist contractures | 1239 | 4 | 6 | 67 | 11 | 18 | 61 | 0 | 3 | 11 | 21 | 52 | 15 | 27 | 56 | ||
| Proximally placement of thumb | 9623 | 5 | 19 | 26 | 7 | 17 | 41 | 0 | 3 | 7 | 20 | 35 | 12 | 39 | 31 | ||
| Metacarpophalangeal joint contractures | 6070 | 2 | 5 | 40 | 10 | 16 | 63 | 0 | 3 | 10 | 19 | 53 | 12 | 24 | 50 | ||
| Camptodactyly | 12385 | 20 | 33 | 61 | 16 | 19 | 84 | 12 | 24 | 50 | 28 | 43 | 65 | 48 | 76 | 63 | |
| Ulnar deviation of any finger | 9465 | 15 | 32 | 47 | 11 | 18 | 61 | 2 | 20 | 10 | 13 | 38 | 34 | 28 | 70 | 40 | |
| Radial deviation of any finger | 9466 | 6 | 9 | 67 | 12 | 18 | 67 | 0 | 3 | 12 | 21 | 57 | 18 | 30 | 60 | ||
| Overlapping toe(s) | 1845 | 4 | 10 | 40 | 4 | 14 | 29 | 0 | 1 | 4 | 15 | 27 | 8 | 25 | 32 | ||
| Proximally placed toes | 1780 | 9 | 21 | 43 | 5 | 14 | 36 | 0 | 3 | 5 | 17 | 29 | 14 | 38 | 37 | ||
| Rocker bottom feet | 1838 | 13 | 18 | 72 | 9 | 13 | 69 | 0 | 12 | 9 | 25 | 36 | 22 | 43 | 51 | ||
| Equinovarus deformity-club feet | 8110 | 34 | 45 | 76 | 14 | 20 | 70 | 5 | 23 | 22 | 19 | 43 | 44 | 53 | 88 | 60 | |
| Achilles tendon contracture | 1771 | 21 | 28 | 75 | 5 | 13 | 38 | 0 | 3 | 5 | 16 | 31 | 26 | 44 | 59 | ||
| Skin, nails, hair | High anterior hairline | 9890 | 14 | 21 | 67 | 8 | 14 | 57 | 0 | 2 | 8 | 16 | 50 | 22 | 37 | 59 | |
| Muscle, soft tissue | Distal muscle weakness | 2460 | 29 | 36 | 81 | 18 | 20 | 90 | 8 | 23 | 35 | 26 | 43 | 60 | 55 | 79 | 70 |
| Distal limb muscle atrophy | 3693 | 15 | 18 | 83 | 13 | 17 | 76 | 0 | 3 | 13 | 20 | 65 | 28 | 38 | 74 | ||
| Edema of the dorsum of hands and feet | 7514 | 8 | 19 | 42 | 8 | 15 | 53 | 2 | 11 | 18 | 10 | 26 | 38 | 18 | 45 | 40 | |
| Neurology | Motor delay | 1270 | 44 | 48 | 92 | 19 | 21 | 90 | 3 | 12 | 25 | 22 | 33 | 67 | 66 | 81 | 81 |
| Inability to walk | 2540 | 15 | 18 | 83 | 17 | 18 | 94 | 0 | 3 | 17 | 21 | 81 | 32 | 39 | 82 | ||
| Generalized hypotonia | 1290 | 15 | 29 | 52 | 7 | 14 | 50 | 1 | 13 | 8 | 8 | 27 | 30 | 23 | 56 | 41 | |
| Intellectual disability | 1249 | 44 | 48 | 92 | 16 | 20 | 80 | 20 | 32 | 63 | 36 | 52 | 69 | 80 | 100 | 80 | |
| Dysarthria, deficit in expressive language | 1260 | 11 | 13 | 85 | 7 | 10 | 70 | 3 | 9 | 33 | 10 | 19 | 53 | 21 | 32 | 66 | |
| Poor speech | 2465 | 24 | 28 | 86 | 11 | 15 | 73 | 2 | 5 | 40 | 13 | 20 | 65 | 37 | 48 | 77 | |
| Absent speech | 1344 | 11 | 15 | 73 | 3 | 16 | 19 | 0 | 5 | 3 | 21 | 14 | 14 | 36 | 39 | ||
| Drooling | 2307 | 26 | 33 | 79 | 8 | 14 | 57 | 1 | 19 | 5 | 9 | 33 | 27 | 35 | 66 | 53 | |
| Dysphagia | 2015 | 9 | 10 | 90 | 8 | 12 | 67 | 0 | 2 | 8 | 14 | 57 | 17 | 24 | 71 | ||
| Chewing difficulties including oral motor dysfunction | 5216 | 12 | 16 | 75 | 6 | 9 | 67 | 2 | 11 | 18 | 8 | 20 | 40 | 20 | 36 | 56 | |
| Spasticity | 1257 | 27 | 35 | 77 | 12 | 17 | 71 | 1 | 15 | 7 | 13 | 32 | 41 | 40 | 67 | 60 | |
| Seizures | 1250 | 21 | 42 | 50 | 4 | 14 | 29 | 2 | 14 | 14 | 6 | 28 | 21 | 27 | 70 | 39 | |
| Delayed CNS myelination | 2188 | 5 | 19 | 26 | 4 | 11 | 36 | 0 | 0 | 4 | 11 | 36 | 9 | 30 | 30 | ||
| Global brain atrophy | 2283 | 8 | 18 | 44 | 3 | 13 | 23 | 0 | 0 | 3 | 13 | 23 | 11 | 31 | 35 | ||
| Ventriculomegaly | 2119 | 3 | 7 | 43 | 5 | 13 | 38 | 0 | 0 | 5 | 13 | 38 | 8 | 20 | 40 | ||
| Hyperreflexia | 1347 | 18 | 26 | 69 | 6 | 9 | 67 | 0 | 11 | 6 | 20 | 30 | 24 | 46 | 52 | ||
| Emotional lability | 712 | 4 | 6 | 67 | 1 | 7 | 14 | 0 | 1 | 1 | 8 | 13 | 5 | 14 | 36 | ||
- Note: ZARD-related clinical features are listed according to the nomenclature/systematics of the OMIM “Clinical synopsis” and were mapped to the Human Phenotype Ontology (HPO). Only positive informative clinical features were scored. Additional less frequent (< 30%) ZARD-related clinical features noticed in this study and reported in the literature are listed in Table S2.
- Abbreviations: %, percentage; HPO, human phenotype ontology; nr, number; n/a, not applicable; w, weeks; y, years.
- *positive informative/total informative.
Prenatal presentation of five fetuses with ZARD (two males and three females), included club foot/feet, rocker bottom feet, fetal hypo/akinesia (mostly at the end of the second or third trimester of pregnancy, > 18–26 weeks of gestation), contractures, AMC, nuchal and/or frontal head edema and (nearly) normal growth parameters (See Videos S1–3 and Figure S3). One affected male fetus presented with hypogenitalism (cryptorchidism and micropenis).
At birth and neonatal age, the clinical genetic diagnosis in males varied from descriptive clinical features, for example, AMC with or without additional clinical features including: Psychomotor developmental delay, short stature, brain atrophy, microcephaly, tetraplegia, congenital general hypotonia, facial weakness-palsy, muscle weakness, and complex spasticity, (flexion) contractures of large and/or small joints, congenital hip dislocation – subluxation, rocker bottom feet, club feet, camptodactyly, and ptosis, to (un)recognizable syndromes, such as cerebral palsy, MCS, WRWF with or without cleft palate, feeding difficulties, laryngomalacia, hyperinsulinemic hypoglycemia, mixed obstructive, and central sleep apnea.
The clinical genetic features in 33 newly identified and published carrier females (from childhood till late adulthood) with a maternally inherited ZC4H2 variant varied from normal phenotype (n = 13) to mild (mainly hand-finger) flexion contractures, borderline to mild ID (n = 20) with facial dysmorphism such as ptosis, strabismus, and long (flat) philtrum. Also, distal muscle weakness, urine (stress) incontinence, anosmia, and walking difficulties which were slowly progressive in a few (n = 3) females in late adulthood were reported.
In marked contrast, the 25 females with a de novo pathogenic variant (from neonatal age till adulthood) of ZC4H2 including Xq11.2 microdeletions showed high variation in clinical presentation and ranged from mildly to severely affected (Table 1; Table S2).
Overall, common clinical features in child- and adulthood in both males and females (30% or more positive informative in probands) included postnatal growth retardation, generalized hypotonia, motor delay, inability to walk, spasticity, hyperreflexia, urinary incontinence, dysarthria-deficit in expressive language, poor or absent speech, ID, drooling, dysphagia, chewing difficulties including oral motor dysfunction, feeding difficulties, facial weakness-palsy, high forehead, high anterior hairline, ocular motor apraxia, strabismus, anteverted nares, microretrognathia, AMC, limited shoulder movement, elbow, wrist contractures, metacarpophalangeal joint contractures, camptodactyly, radial deviation of any finger, knee flexion contractures, equinovarus deformity-club feet, Achilles tendon contracture, distal limb muscle atrophy, or weakness, and micropenis or cryptorchidism in males (Table 1; Table S2, Figures S4 and S5).
Less commonly reported clinical features, which were present in <30% of the probands included short stature, microcephaly (more prominent in males (16/26) than in females (4/30)), round face, low-set (posteriorly rotated) ears, upslanting palpebral fissures, almond-shaped eyes, ptosis, deeply set eyes, pupillary dysfunction, nystagmus, short nose, short philtrum, broad alveolar ridges, high-arched palate, and (submucous) cleft palate (Figures S3 and S4). Furthermore, U-shaped upper lip vermillion/carpshaped mouth and downturned corners of the mouth were reported. A short neck (with limited rotation) was more frequently reported for females with a de novo variant (12/14) than for affected males with an inherited or de novo variant (12/37; Table 1; Table S2, Figure S5). Of note, neonatal respiratory distress, recurrent aspiration pneumonia, and (obstructive sleep) apnea can be a particular concern with apnea more frequently reported in males (11/16) than in affected females with a de novo variant (3/11). In addition reported clinical features included narrow chest, narrow shoulders-thorax (more frequently reported in females with a de novo variant (11/15) than in affected males (18/36); Figure S5), umbilical hernia (Figure S3A, 6.III:1), cervical and/or thoracic kyphosis, scoliosis, narrow pelvis, congenital hip contracture, hip dislocation/subluxation, short limbs, proximally placed thumb, ulnar deviation of any finger, overlapping – proximally placed toe(s), edema of the dorsum of hands and feet (Figure S5), generalized hypotonia, absent speech, epileptic seizures, abnormal cortical gyration, delayed CNS myelination, global brain atrophy, and ventriculomegaly (Figure S6). So far, impaired smell was only reported in carrier females in late adulthood. Upslanting palpebral fissures (Figure S4), arrhythmia, sick sinus syndrome, hypoglycemia, and aggressive behavior were so far only reported in males (Table 1 and Table S2).
Besides the ZARD typical features such as AMC, hypo-/akinesia, club foot/feet and facial dysmorphy, associated sporadic clinical features which can be a clue to clinical diagnosis in probands with ZARD include pupillary dysfunction, optic disk cupping, short neck with limited rotation with narrow chest – thorax – shoulders with limited shoulder movements, malposition of the stomach, diaphragmatic eventration, focal autonomic seizures, electrical status epilepticus during slow sleep (ESES) seizure pattern on electroencephalogram (EEG), pancreatic hypoplasia (and postprandial hypoglycemia), and congenital achalasia (Table S2).
Central and peripheral neurological findings in affected individuals included: ID, poor or absent speech, dysarthria, deficit in expressive language, dysphagia, drooling, and chewing difficulties including oral motor dysfunction, motor delay, inability to walk, spasticity, hyperreflexia, areflexia, and dystonia. Various types of seizures, generalized hypotonia, impaired smell (3/13, adult carrier females), and urinary (stress) incontinence (16/28) were also mentioned.
MRI brain and spine images showed variable and global brain atrophy (11/31; examples are given in Figure S6), delayed CNS myelination (9/30), abnormality of periventricular white matter (4/27), corpus callosum abnormality (7/26), abnormal cortical gyration (5/24), ventriculomegaly (8/20), polymicrogyria, anterior to posterior gradient (1/8 males), tethered cord (3/19), and hydromyelia (2/10 males). Abnormal peripheral nerve conduction was present in one affected (1/7 males). Behavioral phenotypes included emotional lability (5/14) and aggressive behavior (3/5 males; Table S2).
Cardiovascular associated clinical features, for example, arrhythmia (5/13), bradycardia (5/8), (congenital) sick sinus syndrome (5/13), and right ventricular hypertrophy (4/12) were so far reported in affected males only (Table S2).
3.2 ZC4H2 genetic characteristics in the new cohort and global mutational spectrum
Most of the novel families were investigated by WES, WGS and/or array-CGH. For two affected index males (family 4, II:2 and family 6, III:1) it was assumed that the phenotype could be caused by a pathogenic ZC4H2 variant and therefore all coding exons were screened by Sanger sequencing.
A total of eight males from six families (families 1, 3, 4, 5, 9, and 19) inherited the variant from a healthy carrier mother, while three males from three families (families 5, 6, and 9) inherited the variant from a mildly affected mother (Figure 1). For one affected male (family 9, II:5) the phenotype of his mother is unknown. In the moderately affected male from family 18 (Figure 1, II:1 and Figures S4 and S5) and the severely affected male from family 24 (Figure 1, II:1) the missense variant occurred de novo.
In 15 females the variant occurred de novo (Figure 1), including individual II:1 from family 6 who presented with contractures of her metacarpophalangeal finger joints and is otherwise healthy. By contrast, the female fetus of family 3 (Figure 1, II:3) inherited the pathogenic variant from her healthy mother who is a mosaic and carries the variant in about 10% of her cells (Figure S7) and the two mildly affected females from family 9 (Figure 1, II:3 and II:6) carry a maternally inherited missense variant.
A total of 23 variants were identified (Figure 2a,b,c). Thirteen of the 23 were de novo loss-of-function variants (nonsense, splice-site, frameshift, and CNVs) present in affected females and 10 were missense variants identified in affected males which, except for the two de novo variants in affected males (families 18 and 24), were inherited from their mildly affected or asymptomatic carrier mothers. Twenty of the 23 variants were novel including single nucleotide variants (SNVs) and five de novo microdeletions in females. The microdeletions removed the first exon of ZC4H2 but did not affect any adjacent genes. Two missense variants were defined as recurrent. Families 4 and 24 of this study carry a previously reported p.(Arg198Gln) change in an unrelated family (Hirata et al., 2013). Family 18 of this study has a de novo p.(Arg213Trp) variant previously reported in three unrelated families (Hirata et al., 2013; May et al., 2015). Also, in two families in this study (families 5 and 6) the same amino acid (Ala200) was altered. Finally, the proline residue 201 mutated to serine in a published family (Hirata et al., 2013) was mutated to histidine in family 19 of this study.
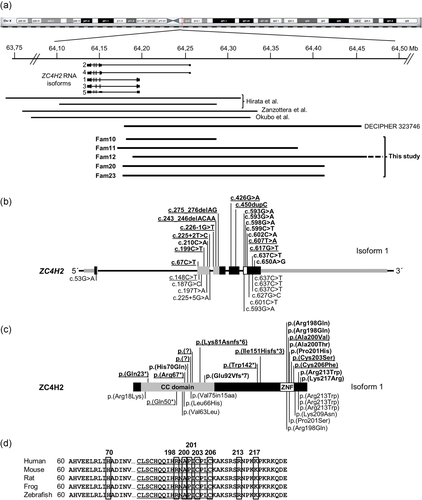
Overview of genetic results in ZARD affected males and females. An overview of newly identified likely pathogenic variants of ZC4H2 described here and previously reported in the literature. (a) Schematic view of the X chromosome with the location of ZC4H2 and the five known RNA isoforms. Horizontal bars indicate the de novo complete ZC4H2 deletions in affected females published previously (Hirata et al., 2013; Okubo et al., 2018; Zanzottera et al., 2017), DECIPHER case 323746 and the newly identified de novo ZC4H2 microdeletions removing part of ZC4H2 identified in this study (families 10–12, 20, and 23). All deletions removed ZC4H2 exon 1 but no other known gene. (b) The structure of human ZC4H2 (Isoform 1; NM_018684.3) with nontranslated sequences gray-striped, exons encoding the coiled-coil domain in gray, and exons encoding the zinc-finger domain in white. Newly identified pathogenic ZC4H2 variants identified in this study are depicted above the gene with de novo variants in affected females underlined. Likely pathogenic variants from the literature are depicted below the gene and protein. (c) Schematic representation of ZC4H2 protein isoform 1 (NP_061154.1) with its functional domains (CC domain: Coiled coil domain; ZNF: zinc-finger domain). ZC4H2 variants newly identified in this study are depicted above the protein with de novo variants in females underlined and variants from the literature are depicted below the protein. (d) Multiple sequence alignment of ZC4H2 protein sequences in five species showing 100% conservation of the newly identified mutated amino acids (boxed). Amino acids of the ZC4H2 zinc-finger domain in the C-terminal part of the protein are underlined
None of the variants identified in this study were reported in publicly available population databases, including the 1000 Genomes Project database and Genome Aggregation Database (gnomAD, http://gnomad.broadinstitute.org/). Also, there were no deletions reported in the database of genomic variants (DGV, http://dgv.tcag.ca/dgv/app/home?ref=GRCh37/hg19) overlapping ZC4H2 exons, demonstrating that the variants identified in the patients are likely not common polymorphisms and supporting our suggestion that they are deleterious to protein function. All missense variants altered highly conserved amino acids (fully conserved from human to zebrafish; Figure 2d and Figure S8). Also, in silico predictions using CADD revealed high scores (> 22) and using Provean all variants were predicted as deleterious, except for the p.(Lys217Arg) change, which perfectly cosegregated with the phenotype in a large family (Fam9). At the gene level, ZC4H2 is highly constrained with a pLI score of 0.91 and zero known LOF mutations in gnomAD (Lek et al., 2016).
Combined with our results of this study, there are currently 31 unique ZC4H2 variants known in affected males and females, including one de novo X-inversion interrupting ZC4H2 in a male plus 10 de novo Xq11.2 microdeletions in females (Figure 2a). An overview of all variants is provided in Table S3. Thus far, SNVs leading to missense changes cluster in the last exon of ZC4H2, which encodes the zinc-finger domain and the most C-terminal part of the protein.
The X-inactivation pattern in blood or skin fibroblasts in affected females varied from random to skewed and was not useful to predict the phenotype (Figure 1 and Supporting Information Results), for example, one severely affected female fetus showed an Xi ratio of 61:39 in skin fibroblasts (Figure 1, Fam3, II:3). Other mild to severely affected females showed Xi ratios varying between 100:0 and 80:20 (Figure 1) in blood lymphocytes. Previous asymptomatic carrier females reported in the literature showed also a ratio of > 95:5 (e.g., Hirata et al., 2013).
3.3 Zebrafish studies
We investigated the potential effects of the p.(Ala200Val) and p.(His70Gln) missense variants which we identified early on in zebrafish morphants similar to our previous studies (Hirata et al., 2013). Following knockdown of zc4h2, zebrafish morphants showed impaired swimming capability at 2 days postfertilization (27/34, 79% of morphants) due to compromised swimming contraction. This defect could be rescued with wild-type Zc4h2 (7/29, 24% of morphants), but not with constructs carrying the p.(Ala200Val) (27/35, 77% of morphants) or p.(His70Gln) (29/41, 71% of morphants) variants and thus the results strongly supported pathogenicity of the altered amino acids.
4 DISCUSSION
This study reports on the genetic and clinical findings of 23 novel families and simplex cases with deleterious inherited or de novo ZC4H2 variants identified in males and females, including de novo partial ZC4H2 microdeletions present in affected females only, and a review of the ZARD literature. Moreover, we have investigated pathogenicity of two single amino acid substitutions in zebrafish as described before (Hirata et al., 2013).
Inherited and de novo variants are present in all coding exons and there is no hotspot in the gene. On the protein level, most of the missense changes lie within the C-terminal part of ZC4H2 including the zinc-finger domain. All mutated amino acids of ZC4H2 are highly conserved and predicted to be functionally relevant. The two missense variants tested in zebrafish resulted in impaired swimming of the mutants, supporting functional importance of the mutated amino acids. Two of the de novo pathogenic missense variants identified in females (families 8 and 15, p.(Cys206Phe) and p.(Cys203Ser) respectively) altered one of the four cysteine residues of the Cys4His2 type zinc-finger. The function of this zinc finger is currently unknown.
Most ZC4H2 pathogenic variants identified in affected males are missense changes and, except for two de novo missense variants identified in affected males (families 18 and 24), were inherited from mostly asymptomatic mothers (X-recessive). This is well in line with previous findings. To the best of our knowledge, with only two exceptions there are no pathogenic variants reported in affected males predicted to lead to a truncated or completely absent ZC4H2 protein. The two exceptions are the de novo X-chromosome inversion which disrupted ZC4H2 at one of the breakpoints in a severely affected boy (Hirata et al., 2013) and the affected boy from family 3 who we assume, based on clinical presentation, carried the maternally inherited p.(Glu92Vfs*7) variant but did not undergo genetic testing. He was born after Caesarean section because of fetal distress at 40 weeks pregnancy with multiple congenital malformations and died 10 hr after birth because of respiratory distress. He had short limbs, AMC and a cleft palate. Affected males can have syndromic variable XLID (mild-moderate ID) with (mild) hypotonia, spasticity (as seen in SPG16 [Steinmuller et al., 1997]), mild facial dysmorphism, for example, high forehead with high frontal hairline, ocular dyspraxia, relative short stature, and (relative) microcephaly. The differential diagnosis is still challenging and earlier reported XLID families could benefit from WES including ZC4H2 analysis (Table S4).
In contrast to the mostly inherited missense variants in affected males, the spectrum of de novo pathogenic ZC4H2 variants identified in affected females includes missense, early stop, frameshift and splicing variants, as well as Xq11.2 microdeletions which removed ZC4H2 completely or exon 1 of all transcript isoforms. Thus, from the current data it seems that pathogenic variants predicted to lead to a complete loss of ZC4H2 protein function are very rare in males, while they usually occur de novo in females. Of note, none of the pathogenic variants identified in males were found de novo in female simplex cases. The de novo variants in the affected females are predicted to be loss-of function alleles, suggesting ZC4H2 insufficiency as the most likely pathological mechanism leading to an X-linked dominant phenotype.
Our detailed clinical characterization (Table 1 and Table S2) confirms and extends clinical findings reported for the few published families and simplex cases caused by deleterious ZC4H2 SNVs in males and de novo ZC4H2 microdeletions in females. Furthermore, we report the first mosaic frameshift variant of ZC4H2 in a family with AMC and fetal hypo-/akinesia. Combined, the results indicate that, in males, inherited forms of ZARD are associated with complete penetrance but variable expressivity, ranging from nonspecific XLID to XL-AMC with variable mostly moderate to severe forms of ID, fetal hypo-/akinesia, postnatal growth retardation, spasticity, tetraplegia, (relative) microcephaly, and hypogenitalism. By contrast, in females, the inherited forms of ZARD are associated with incomplete penetrance and variable expressivity, ranging from unaffected carriers to XL-AMC with variable cognition, postnatal growth retardation, poor or absent speech, spasticity, inability to walk, distal muscle wasting/atrophy, short neck with limited rotation, narrow chest, and limited shoulder movements. Moreover, clinical follow-up of family 7 for more than 25 years (Hennekam et al., 1991) indicated very mild progressive muscle weakness in two former asymptomatic carrier females and marked progression including loss of the ability to walk and distal muscle wasting in one carrier female (Figure S2, III:5, III:7, and III:14). She also had episodic periods of pain of unknown origin. All three females had inability to smell and urine stress incontinence. As differential diagnoses a late-onset mild progressive syndromic form of spastic paraplegia (SPG), multiple sclerosis (MS), amyotrophic lateral sclerosis (ALS), or Charcot-Marie-Tooth (CMT) were clinically (re)considered. While we cannot exclude comorbidity with other X-linked forms of SPG (e.g., PLP1, SLC16A2, and TAF1), ALS (SMAX1), and CMT (GJB1) which could not be tested genetically, the females are all carriers of a ZC4H2 pathogenic variant causing ZARD in their male offspring. This makes it more likely that their late-onset phenotype belongs to ZARD's clinical variability. In contrast to this late-onset neurodegenerative adult phenotype reported so far in one family, there is no early-onset progressive neurodegenerative phenotype known in affected children.
Compared with males, females with a pathogenic de novo variant of ZC4H2 presented with a more variable phenotype. Cognition ranged from normal (families 3, 6, and 23) to mild learning difficulties (families 8, 21, and 22), as reported for affected who carry missense, frameshift or splicing variants, and a partial gene deletion, respectively, to moderate or severe ID (families 10–17). Furthermore, clinical characteristics of the de novo Xq11.2 microdeletions present in females showed that these deletions are associated with a recognizable phenotype and that the overall clinical outcome varies between mildly and severely affected and thus cannot be predicted. Speech delay and poor speech are a remarkable clinical feature in the affected, so speech therapy should be started as early as possible to optimize communication leading to improved quality of life. Also, a large Xq11.2 deletion which, in addition to ZC4H2, removed ARHGEF9, WTX, and MTMR8 was reported in a female with severe ID and normal X-inactivation (Holman et al., 2013). The major clinical phenotype was caused by the WTX deletion leading to osteopathia striata with cranial sclerosis and it was suggested that deletion of ZC4H2 contributed to her severe ID. Given the results obtained through this study it is possible that in addition to ID other clinical features reported for this female, such as clenched hands with immobile joints, early-onset complex partial seizures (controlled on levetiracetam), dysphagia, and lower extremity spasticity could be caused by ZARD.
Two of the pathogenic ZC4H2 missense variants identified in this study are recurrent. First, the maternally inherited amino acid change p.(Arg198Gln) identified in the affected males of one family (family 4) and de novo in the affected male of an unrelated family (family 24) has previously been reported in a large German family (Hirata et al., 2013) and there are phenotypic overlaps with affected males being severely affected. Secondly, the de novo p.(Arg213Trp) substitution identified in a moderately affected male (family 18) has previously been reported in three large families (Hirata et al., 2013; May et al., 2015). The clinical phenotype is variable within and between families, but all affected males (Family K8615, May et al., 2015, families 4 and 5, Hirata et al., 2013) presented with moderate to severe ID with absent or poor speech. Mild facial dysmorphism was evident in one family only (family 5, Hirata et al., 2013). Other common shared clinical features included motor delay, inability to walk, hyperreflexia, spasticity, and seizures. Skeletal findings, for example, AMC were reported in a few affected males (2/2 males of family 4 and 1/2 males of family 5, Hirata et al., 2013). All female carriers, except one, of the three families had borderline or mild ID (family K8615, May et al., 2015, families 4 and 5 Hirata et al., 2013).
Overall, despite the growing number of families and simplex cases with deleterious ZC4H2 variants, there is currently no evidence for a clear genotype-phenotype correlation. Also, as discussed above, clinical presentations of affected individuals who carry the same pathogenic variant leading to ZARD can vary within families and between families. Nevertheless, there are some gender-specific clinical features, for example, heart rhythm disturbances and hypogenitalism in males.
The striking phenotypic differences between females with inherited X-linked recessive ZC4H2 missense variants versus de novo X-linked dominant deleterious ZC4H2 nonsense, frameshifting or Xq11.2 microdeletions is similar to that seen for a few other X-linked genes, such as ARX (Bienvenu et al., 2002; Mattiske et al., 2017; Stromme et al., 2002), HDAC8 (Kaiser et al., 2014), PHF6 (Lower et al., 2002; Zweier et al., 2013), IQSEC2 (Ewans et al., 2017; O'Rawe et al., 2015; Shoubridge et al., 2010; Zerem et al., 2016), and CLCN4 (Hu et al., 2016; Palmer et al., 2018). Also gender-specific pathogenicity differences of inherited variants in males and de novo variants in females have been reported for other XLID genes, for example, DDX3X (Dikow et al., 2017; Snijders Blok et al., 2015), KIAA2022 (Lorenzo et al., 2018; Van Maldergem et al., 2013), MTM1 (Schara, Kress, Tucke, & Mortier, 2003), and CASK (Moog et al., 2011).
ZC4H2 is known as a gene which is subject to X-inactivation. Current results suggest that, in heterozygous carrier females, X-inactivation status in blood and skin fibroblasts does not predict the clinical outcome. This is very much in line with results obtained for other X-linked disease genes and also with recent findings suggesting that skewed X-inactivation is common in the general female population (Shvetsova et al., 2018). Still, the variable clinical manifestations of heterozygous carrier females with a de novo variant ranging from very mildly to severely affected may be partially explained by the X-inactivation status within specific affected cells and tissues. However, other genetic, environmental, or stochastic factors may also be involved and could have an impact on phenotypic variability and severity of ZARD.
Similar to patients with other types of AMC who develop joint contractures during pregnancy, abnormal fetal movement due to ZARD can be identified using real time ultrasound prenatally, as we report here for affected fetuses from several families (for family 3 see Videos S1–3). An early diagnosis of AMC prenatally would allow parents and clinicians early decision making, for example, the possibility of in utero therapy (increasing movement in utero), or early delivery at a time when lungs are mature but the contractures may not yet be so severe (Hall, Agranovich, Ponten, & van Bosse, 2015). However, AMC and fetal hypo-/akinesia can occur late in pregnancy, after routine prenatal ultrasound sonography has taken place between 18 and 22 weeks of gestation, thus the diagnosis of AMC and/or fetal hypo-/akinesia can be easily missed. Moreover, reduced fetal hand, finger and feet movements are not examined routinely by ultrasound and occasionally club foot/feet is/are the only clinical feature which fetuses with a pathogenic ZC4H2 variant presented prenatally or neonatally.
In conclusion, ZC4H2 is one of the more commonly mutated XL-AMC and XLID genes. Up to now de novo pathogenic variants of ZC4H2 have been more frequently seen in affected females than in males with ZARD and displayed a broad clinical spectrum ranging from mildly to severely affected patients with neurogenic AMC with or without CNS and PNS involvement (see Supporting Information). Our findings suggest including ZC4H2 analysis retrospectively in AMC male and female cohorts and prospectively in prenatal and/or neonatal genetic diagnostic tests in male and female fetuses presenting with fetal hypo-/akinesia and/or AMC, for example, (only) club foot/feet with or without hypogenitalism in males. Long-term prospective clinical studies assessing the development of the ZARD phenotypes and investigations determining the molecular and cellular mechanisms underlying the disorders are required.
ACKNOWLEDGMENTS
The authors would like to thank the individuals and their families who participated in this study. We also thank Andrew Green, Christina Fagerberg, and Melissa Lees for giving permission to include publicly available DECIPHER entries on their patients. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003), a parallel funding partnership between Wellcome and the Department of Health, and the Wellcome Sanger Institute (grant number WT098051). The views expressed in this publication are those of the author(s) and not necessarily those of Wellcome or the Department of Health. The study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12 granted by the Republic of Ireland REC). This study makes use of data generated by the DECIPHER community. A full list of centres who contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via email from [email protected]. Funding for the project was provided by the Wellcome Trust. For one of the families this study was made possible through access to the data and findings generated by the 100,000 Genomes Project. The 100,000 Genomes Project is managed by Genomics England Limited (a wholly owned company of the Department of Health). The 100,000 Genomes Project is funded by the National Institute for Health Research and NHS England. The Wellcome Trust, Cancer Research UK and the Medical Research Council have also funded research infrastructure. The 100,000 Genomes Project uses data provided by patients and collected by the National Health Service as part of their care and support. Part of this study was supported by a Dutch NWO VENI grant (OND1312421 to S.F.), NEI grant R01EY027421, and NHLBI grant X01HL132377 (to ECE). The Broad Center for Mendelian Genomics (UM1 HG008900) is funded by the National Human Genome Research Institute with supplemental funding provided by the National Heart, Lung, and Blood Institute under the Trans-Omics for Precision Medicine (TOPMed) program and the National Eye Institute.




