Increasing phenotypic annotation improves the diagnostic rate of exome sequencing in a rare neuromuscular disorder
Abstract
Phenotype-based filtering and prioritization contribute to the interpretation of genetic variants detected in exome sequencing. However, it is currently unclear how extensive this phenotypic annotation should be. In this study, we compare methods for incorporating phenotype into the interpretation process and assess the extent to which phenotypic annotation aids prioritization of the correct variant. Using a cohort of 29 patients with congenital myasthenic syndromes with causative variants in known or newly discovered disease genes, exome data and the Human Phenotype Ontology (HPO)-coded phenotypic profiles, we show that gene-list filters created from phenotypic annotations perform similarly to curated disease-gene virtual panels. We use Exomiser, a prioritization tool incorporating phenotypic comparisons, to rank candidate variants while varying phenotypic annotation. Analyzing 3,712 combinations, we show that increasing phenotypic annotation improved prioritization of the causative variant, from 62% ranked first on variant alone to 90% with seven HPO annotations. We conclude that any HPO-based phenotypic annotation aids variant discovery and that annotation with over five terms is recommended in our context. Although focused on a constrained cohort, this provides real-world validation of the utility of phenotypic annotation for variant prioritization. Further research is needed to extend this concept to other diseases and more diverse cohorts.
1 INTRODUCTION
Next-generation sequencing (NGS) has progressively transformed the way rare diseases are diagnosed from a serial gene-by-gene approach to one in which variation across the whole genome or its protein-coding sequence (exome) is analyzed following a single experimental assay. However, each exome may contain as many as 30,000 variants from the reference sequence (Gilissen, Hoischen, Brunner, & Veltman, 2012), several thousand of which may have a putative protein-altering effect. Despite the growth and increasing sophistication of bioinformatics tools predicting the pathogenicity of these variants based on functional consequences and population frequency alone (Eilbeck, Quinlan, & Yandell, 2017), a typical exome sequence may have as many as 500 rare variants (population frequency <1%) with a potential functional effect (Kernohan et al., 2018). For many rare diseases it therefore remains the case that phenotypic assessment is essential to differentiate between a number of candidate variants that look equally plausible from a genetic perspective when the whole exome or genome is evaluated. Deep phenotyping of undiagnosed cases thus remains a crucial part of diagnosis and gene discovery in rare disease (Boycott et al., 2017), but the subsequent variant assessment may still rely on a disease expert's knowledge or recognition of a specific phenotype rather than on computer guidance. We aimed to test the hypothesis that increased phenotypic annotation using the Human Phenotype Ontology (HPO; Köhler et al., 2019) as a standardized terminology system aids computer-based detection and prioritization of the correct causative variant.
Clinicians submitting whole-exome and whole-genome sequencing data (WES/WGS) to national and international diagnostic or gene discovery initiatives such as RD-Connect, the Undiagnosed Diseases Network International, the US Centers for Mendelian Genomics or the UK 100,000 Genomes Project are required to provide phenotypic annotations using the HPO for each case they submit (Köhler et al., 2019). The HPO provides a highly comprehensive and granular set of terms for annotating individual atomic phenotypes observed in the patient within a logical hierarchical structure and has become the most widely used phenotypic annotation mechanism across the rare disease field for diagnosis and gene identification. Various mechanisms are then available for incorporating this phenotypic knowledge into the diagnostic or discovery workflow. At the most straightforward level, it is possible to restrict searches to “virtual panels” of genes associated with a particular phenotype. The best known of these approaches mimics the diagnostic approach of sequencing using gene panels by filtering an exome or genome against a curated shortlist of genes that have already been implicated with a specific global phenotype such as NMD (a disease-gene list). As an alternative which has been made possible thanks to gene:HPO associations curated from the literature and online databases and available from the HPO developers, it is possible to use individual atomic phenotypes to build a gene list on the fly that collates all genes associated with any of the individual HPO terms. Both of these gene list approaches provide a simple yes/no, presence/absence result. More sophisticated algorithms are also available, such as those used by the Exomiser suite of tools (Smedley et al., 2015), some of which use comparisons from animal models as well as human data (Bone et al., 2016). These enable computer-aided assessment and numerical ranking of the likelihood that a particular variant is causative.
However, information about the depth to which phenotypic annotation should be done to maximize the chance of finding the causative variant is largely lacking. During development of the algorithms, researchers created simulations in which they spiked unaffected exomes with causative variants and trialed them with synthetic HPO term sets. Other work has been done through the Monarch Initiative to develop methods for assessing “phenotypic annotation sufficiency” and provide metrics to assist users (Washington et al., 2014) and some phenotypic annotation tools such as PhenoTips incorporate these methods to help submitters assess how complete their annotation is (Buske, Girdea et al., 2015). More analyses on real datasets are of benefit to confirm these tests in a real-world situation.
The RD-Connect Genome Phenome Analysis Platform (GPAP) is an international database and analysis interface for genomic and phenotypic data that contains whole-exome and whole-genome data and corresponding phenotypic information from over 5,000 individuals with rare disease and family members (Lochmüller et al., 2018). The RD-Connect platform provides an opportunity to test phenotypic analysis algorithms on real data. A preliminary analysis of a subset of cases (n = 210) in the platform from the BBMRI-LPC 2016 whole-exome sequencing call for rare diseases (http://www.bbmri-lpc.org/) showed a linear correlation between the number of individual HPO terms each case has been annotated with and the proportion of cases that have received a solution in the platform, with the greater the number of HPO annotations, the more likely the case is to have been solved (Beltran, S. unpublished data). Recent data on the diagnosis rate coming out of the 100,000 Genomes Project has shown a similar result (Caulfield, M., 2018). However, such analyses contain many potentially confounding variables, including differences in typical annotation levels and in solve rates for different disease domains, as well as the consideration that cases that have been more extensively studied by the submitter may have both more comprehensive annotation and also a greater likelihood of being solved without their being a causal relationship between the two. Without further investigation, it is therefore difficult to consider the correlation as proof that increasing phenotypic annotation is in itself the cause of the improved solve rate.
Our experimental setup was designed to answer a number of related questions using real data but in a controlled setting. How well does a traditional disease-gene filter (equivalent to a diagnostic panel) perform in capturing the causative variant while excluding nonrelevant variants? How does the performance of a gene list derived from HPO annotations compare to an expert-curated disease-gene list? How well does Exomiser perform against the gene-list strategies when using the human data comparison algorithm; what added value does the human, animal and interactome comparison provide; and what is the benefit of adding more phenotypic annotations? To answer these questions, we set up two sets of experiments using the same patient cohort. As described in Section 2 and depicted in Figure 1, experiment set 1 focused on gene list filters, while experiment set 2 focused on the Exomiser tool for variant prioritization. Experiment 1a filtered variants against a published disease-gene list for neuromuscular disorders (Kaplan & Hamroun, 2015). Experiment 1b filtered the same variants against a list of genes generated from a standardized set of clinically relevant HPO terms, while Experiment 1c filtered them against individual gene lists derived from the HPO terms entered by the submitting clinician. Experiment 2a used the Exomiser tool with its human data comparison algorithm to prioritize variants based on different numbers of standardized and submitter-entered HPO terms and assess the added value of increased numbers of annotations, while Experiment 2b performed the same tests using the human, model organism and interactome comparison algorithm to assess the relative performance of the two algorithms on our cohort.
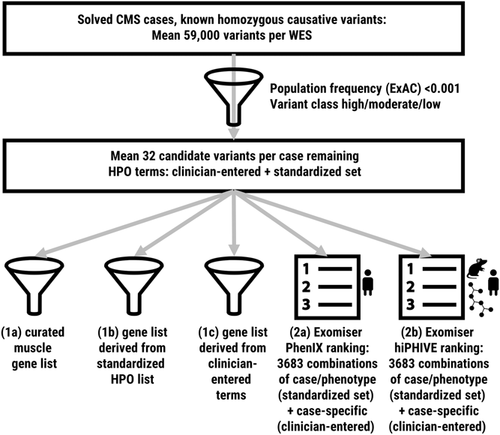
Experimental setup. We performed five analyses on the same cohort of 29 patients using both a standardized set of HPO terms and the clinician-entered terms unique to each case. We first assessed the detection rate of gene list filters and then assessed the performance of Exomiser with Phenix and hiPHIVE algorithms. CMS, congenital myasthenic syndrome; HPO, Human Phenotype Ontology; WES, whole-exome sequencing
2 MATERIALS AND METHODS
2.1 Editorial policies and ethical considerations
The RD-Connect GPAP has received ethics approval from the Parc de Salut MAR Clinical Research Ethics Committee on 27 October 2015 under Ref. No. 2015/6456/I. All data used in this study had been originally submitted to RD-Connect under the ownership of the study principal investigator (PI), H. L., and was accessed by authorized researchers in compliance with the RD-Connect Code of Conduct. As with all data in the RD-Connect GPAP, no individuals in this study were personally identifiable and all had been consented by the submitting clinician to analysis of their data for research purposes in accordance with local ethics regulations.
2.2 Patient cohort
Since our aim was to explore the effect of varying phenotypic annotation on the solve rate in a real patient cohort, we needed to select a set of patients whose cause of disease was already known to establish our ground truth, and to minimize nonphenotypic variables to focus on the effect of phenotype. From the cases available within the study PI's cohort in the RD-Connect GPAP, we selected a restricted cohort of 29 individuals with a confirmed genetic diagnosis (“solved cases”), all of which had a similar phenotypic profile and homozygous causative variant. All individuals had originally been clinically suspected of a congenital myasthenic syndrome (CMS; Thompson et al., 2018) based on their phenotypic presentation to the submitting clinician and had been submitted within the RD-Connect PhenoTips instance as CMS cases. All were subsequently diagnosed with a homozygous recessive cause of disease that was confirmed by further analysis by the submitting center. Our review of the cases confirmed the compatibility of the phenotypic description with CMS. The aim of restricting the initial analysis cohort in this way was to minimize the effect of the genetic variation on the analysis to focus on the effect of phenotype on the analysis results. The 29 cases covered 18 different genes in total (see Table 1). Twenty two of the twenty nine cases (11 genes) had causative variants in genes that have been previously associated with CMS according to the Online Mendelian Inheritance in Man (OMIM) database (Amberger, Bocchini, Scott, & Hamosh, 2019); 5 cases (5 genes) had variants in genes that were associated with other human disease but not CMS; and 2 cases (2 genes) had no human disease associated according to OMIM but have been assigned as causative by the submitting clinician on the basis of segregation and downstream analysis. The exomes analyzed came from several different projects and were not all from the same original sequencing provider but had all been reprocessed from the raw data (FASTQ or untrimmed BAM) using the standard RD-Connect analysis pipeline (Laurie et al., 2016) to standardize variant calling and annotation in line with standard RD-Connect practice.
| Genes | No. cases | OMIM disease associations | Comment |
|---|---|---|---|
| AGRN | 1 | CMS | Well known CMS gene |
| CHAT | 1 | CMS | Well known CMS gene |
| CHRND | 1 | CMS | Well known CMS gene |
| CHRNE | 2 | CMS | Well known CMS gene |
| COLQ | 4 | CMS | Well known CMS gene |
| DOK7 | 1 | CMS | Well known CMS gene |
| GFPT1 | 5 | CMS | Well known CMS gene |
| MUSK | 3 | CMS | Well known CMS gene |
| COL13A1 | 1 | CMS | Recent CMS gene—annotated in OMIM but no phenotype result from Exomiser and therefore considered a novel gene for the purposes of this study |
| DPAGT1 | 2 | CMS + congenital disorder of glycosylation, type Ij | Known CMS gene but also CDG-associated |
| SLC25A1 | 1 | CMS + combined d-2- and l-2-hydroxyglutaric aciduria | Recent CMS gene—annotated in OMIM but considered a novel gene for the purposes of this study |
| CLP1 | 1 | Pontocerebellar hypoplasia type 10 | No CMS association in OMIM. Some phenotypic overlap |
| DHCR7 | 1 | Smith-Lemli-Opitz syndrome | No CMS association in OMIM. Some phenotypic overlap |
| HSPG2 | 1 | Schwartz-Jampel syndrome type 1 | No CMS association in OMIM. Some phenotypic overlap |
| POLG | 1 | Progressive external ophthalmoplegia, mitochondrial DNA depletion syndrome, mitochondrial recessive ataxia syndrome | No CMS association in OMIM. Some phenotypic overlap |
| TOR1A | 1 | Dystonia-1, torsion | Also recently associated with arthrogryposis multiplex congenita |
| CTNND2 | 1 | None | Candidate gene |
| TENM2 | 1 | None | Candidate gene |
- Abbreviations: CDG, congenital disorders of glycosylation; CMS, congenital myasthenic syndrome; OMIM, Online Mendelian Inheritance in Man.
Knowing the confirmed diagnosis of these cases allowed us to assess the success of different analyses in terms of their ability to prioritize the correct causative variant. A standard workflow recreating the recommended analysis approach for a new case in RD-Connect was applied to all cases. First, the total number of variants in each exome before applying filters was assessed. Then, to reduce the number of variants to evaluate in the phenotypic part of the analysis, each exome was filtered using a standard set of genomic filters available within RD-Connect, including variant class and population frequency (Table S1). At each stage it was confirmed that the confirmed pathogenic variant was still present in the resulting data set and had not been filtered out. On achieving a filtered data set of rare, plausibly pathogenic variants based on variant data alone, we applied several methods of prioritizing by phenotype, as described in Experiments 1 and 2 below.
2.3 Standardized HPO set
All individuals selected for the analysis had varying numbers of HPO terms entered by the submitting clinician at the time of submission (ranging from 1 to 18 terms, all specific to that individual), and these were used as part of our analyses as described below. In addition to these submitter-entered HPO terms, we also generated a standardized or simulated set of seven HPO terms using phenotypic features that are common to many of the CMS types and frequently used to describe CMS patients. This standardized set was designed to be applied across all cases to enable us to assess the effect of varying the phenotypic annotation. In this standardized set we endeavored to cover a range of different branches of the HPO hierarchy, including neurological, muscular, respiratory, joint, and eye phenotypes. It was intentionally generated by expert knowledge without reference to the existing HPO gene:phenotype annotations in order not to be biased towards known annotations. We applied the standardized list in Experiments 1b and 2 below. An overview of the experimental workflow is provided in Figure 1.
2.4 Hierarchy of terms and information content
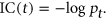
Using disease and annotation numbers available in the online HPO browser (https://hpo.jax.org/app/browse/) we calculated the IC for each of our seven terms in the standardized set. To assess the performance of the algorithms with more general terms with lower information content, we then extracted the parent terms and grandparent terms and calculated the IC in the same way. The values are shown in Table 2.
| Original term set | Parent term set | Grandparent term set | ||||||
|---|---|---|---|---|---|---|---|---|
| HPO ID | Term | IC | HPO ID | Term | IC | HPO ID | Term | IC |
| HP:0100285 | EMG: impaired neuromuscular transmission | 2.83 | HP:0003457 | EMG abnormality | 1.66 | HP:0011804 | Abnormality of muscle physiology | 0.55 |
| HP:0002872 | Apneic episodes precipitated by illness, fatigue, stress | 3.33 | HP:0002104 | Apnea | 1.79 | HP:0002793 | Abnormal pattern of respiration | 1.66 |
| HP:0000597 | Ophthalmoparesis | 1.73 | HP:0000496 | Abnormality of eye movement | 0.80 | HP:0012373 | Abnormal eye physiology | 0.54 |
| HP:0000508 | Ptosis | 1.24 | HP:0012373 | Abnormal eye physiology | 0.54 | HP:0000478 | Abnormality of the eye | 0.38 |
| HP:0001290 | Generalized hypotonia | 1.11 | HP:0001252 | Muscular hypotonia | 0.82 | HP:0003808 | Abnormal muscle tone | 0.68 |
| HP:0003473 | Fatigable weakness | 2.39 | HP:0001324 | Muscle weakness | 0.99 | HP:0011804 | Abnormality of muscle physiology | 0.55 |
| HP:0002804 | Arthrogryposis multiplex congenita | 1.94 | HP:0002803 | Congenital contracture | 1.86 | HP:0001371 | Flexion contracture | 1.11 |
- Note: The original standardized set of HPO terms selected for relevance across CMS cases is presented with the IC value for each term generated from values for annotation and disease numbers available from the HPO website (values downloaded 17 April, 2019). “Parent” and “grandparent” terms were obtained by going up one and two levels respectively in the HPO hierarchy and IC values generated in the same way.
- Abbreviations: CMS, congenital myasthenic syndrome; EMG, electromyogram; HPO, Human Phenotype Ontology; IC, information content.
2.5 Experiment 1a: Disease-gene list
A disease-gene list or “virtual panel” is commonly used in clinical diagnostics where exome sequencing has been performed. This restricts the output to variants in a predefined set of genes that have already been associated with human disease with a particular global phenotypic presentation. Several lists curated by disease domain experts are available within the RD-Connect system and can be applied as a filter on the data. Since our cohort all had a clinical starting diagnosis of CMS, which is a neuromuscular disease (NMD), we applied the 2016 version of the Neuromuscular Gene Table list (Kaplan & Hamroun, 2015), a comprehensive and carefully curated set of genes associated with NMD which has become the de facto standard for the neuromuscular field. To evaluate this filtering method, we defined our detection rate as the proportion of cases in which the known causative variant was among the variants returned. We also assessed the total number of variants returned.
2.6 Experiments 1b and 1c: Gene lists generated from HPO terms
As an alternative to a virtual panel for a disease domain, it is possible to generate a gene list by compiling all genes that have been annotated in the literature as associated with an individual HPO term or collection of terms. This makes use of curated gene:phenotype annotations compiled by the HPO developers and updated on a monthly basis. A PhenoTips-developed RESTful API (The PhenoTips developers, 2017) enables the compilation of a list of genes based on a given set of HPO terms. Our analysis used the annotation set from September, 2018 (The HPO developers, 2018). Here, we performed two analyses. First, we took the actual phenotypic terms entered by the submitting clinician and used the API within RD-Connect to generate a gene list from those terms. Since each case had a unique annotation set, this created a unique gene list for each patient. Second, we used the same RD-Connect function with our seven standardized HPO terms, which enabled us to apply a consistent set of terms and therefore a consistent gene list across the 29 cases. Again our detection rate for both cases was the proportion of cases in which the known causative variant was returned.
2.7 Experiment 2. Variant prioritization using the Exomiser
The Exomiser is an application designed to find and prioritize potential disease-causing variants in WES and WGS data (Smedley et al., 2015). As input it takes a VCF file and a set of phenotypes in HPO, and from this it annotates, filters and prioritizes likely causative variants. It creates a variant score from predicted pathogenicity and population frequency data and a phenotypic relevance score based on a comparison of how closely the given phenotype matches the known phenotype of disease genes. The two scores combined are then used to rank the candidate variants in terms of the likelihood that each may be causative. The phenotypic relevance score can be set to base its results on human data only (PhenIX algorithm), model organism data only (PHIVE), or human, model organism and interactome data combined (hiPHIVE). Mode of inheritance may be entered to include only those results compatible with the assumed inheritance pattern. By providing a score and a ranking for each variant with each phenotypic combination, the Exomiser results provide a more sophisticated method for evaluating the effect of phenotypic annotation beyond the inclusion/exclusion method provided by the gene list experimental setup.
In the RD-Connect GPAP, users have the possibility to submit candidate variants and associated phenotypes to Exomiser for analysis, but it can only be run with the HPO terms entered by the submitting clinician, and not with our standardized set of terms. For this test, therefore, we installed a separate Exomiser instance on a local server. We extracted the filtered candidate variants from the RD-Connect platform in VCF format and used them in Experiments 2a and 2b as follows.
2.8 Experiment 2a: Variant prioritization using the Exomiser with its human data comparison algorithm (PhenIX)
Using the VCFs containing the filtered variants together with the standardized HPO set described above, we set Exomiser to its human-only algorithm (PhenIX) and homozygous recessive inheritance pattern and sequentially ran all possible variations of cases and HPO terms, from one HPO term to seven terms in the standardized set in combination with all 29 cases (3,683 separate combinations). We then repeated the test with the “real” HPO set for each case as entered by the submitter (29 further experiments). We captured the output for the known causative variant, including the gene-variant score (genomic data alone; no consideration of phenotype), gene-pheno score (the match of the phenotype with the gene at gene level, excluding variant pathogenicity assessment), and gene-combined score (combines the variant and phenotype scores to give an overall assessment of likelihood that the variant is causative). We also captured the rank of the causative variant in each case.
To assess whether the level of specificity of our annotations affected the score and rank obtained using the Exomiser algorithm, we also repeated the analysis using the parent and grandparent terms, capturing the same scores and ranks for comparison against those obtained using our original annotations.
2.9 Experiment 2b: Variant prioritization using the Exomiser with its human, model organism and interactome comparison algorithm (hiPHIVE)
We repeated the same experimental setup as for 2a, the only difference being that Exomiser was set to run with the hiPHIVE algorithm. The same set of results was captured and compared against the data for the PhenIX algorithm to provide an assessment of whether and under what circumstances it is beneficial to include comparison against model organism and interactome data.
3 RESULTS
We analyzed the 29 cases using the methods described (see Figure 1 for summary). An overview of the number of variants per exome and the results of the experiments is provided in Table 3. The detection rates of the gene list filters are summarized in Figure 2. The results using the Exomiser algorithms with different HPO combinations are then further analyzed separately.
| Experiment 1a: NMD gene table filter | Experiment 1b: HPO filter, standardized HPO list | Experiment 1c: HPO filter, clinician-entered HPO list | Experiment 2a: Exomiser PhenIX algorithm | Experiment 2b: Exomiser hiPHIVE algorithm | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RD-Connect case No. | Confirmed causative gene | Number of homozygous variants before filtering | Correct variant present after variant prefiltering | Number of variants after variant prefiltering | Correct variant present | Total number of variants | Correct variant present | Total number of variants | Correct variant present | Total number of variants | Rank of correct variant with no HPO used (Exomiser variant score rank) | Rank of correct variant with 7 HPO terms (Exomiser combined rank (standardized HPO list) | Combined rank (clinician-entered HPO list) | Combined rank (standardized HPO list) | Combined rank (clinician-entered HPO list) |
| E425745 | AGRN | 39200 | ✓ | 10 | ✓ | 2 | ✓ | 2 | ✓ | 2 | 1 | 1 | 1 | 1 | 1 |
| E605413 | CHAT | 42697 | ✓ | 44 | ✓ | 2 | ✓ | 3 | ✓ | 5 | 1 | 1 | 1 | 1 | 1 |
| E171200 | CHRND | 65446 | ✓ | 27 | ✓ | 2 | ✓ | 2 | ✓ | 2 | 4 | 1 | 1 | 1 | 1 |
| E578399 | CHRNE | 38494 | ✓ | 22 | ✓ | 3 | ✓ | 3 | ✓ | 3 | 1 | 1 | 1 | 1 | 1 |
| E975942 | CHRNE | 36170 | ✓ | 8 | ✓ | 1 | ✓ | 1 | ✓ | 1 | 1 | 1 | 1 | 1 | 1 |
| E391167 | COLQ | 66235 | ✓ | 6 | ✓ | 1 | ✓ | 1 | ✓ | 1 | 1 | 1 | 1 | 1 | 1 |
| E511806 | COLQ | 78689 | ✓ | 56 | ✓ | 3 | ✓ | 1 | ✓ | 2 | 1 | 1 | 1 | 1 | 1 |
| E615835 | COLQ | 68395 | ✓ | 35 | ✓ | 3 | ✓ | 4 | ✓ | 4 | 1 | 1 | 1 | 1 | 1 |
| E998239 | COLQ | 67769 | ✓ | 28 | ✓ | 2 | ✓ | 3 | ✓ | 2 | 1 | 1 | 1 | 1 | 1 |
| E000053 | DOK7 | 52784 | ✓ | 27 | ✓ | 3 | ✓ | 2 | ✓ | 2 | 1 | 1 | 1 | 1 | 1 |
| E200102 | GFPT1 | 75924 | ✓ | 41 | ✓ | 4 | ✓ | 2 | ✓ | 1 | 2 | 1 | 1 | 1 | 1 |
| E280132 | GFPT1 | 73896 | ✓ | 21 | ✓ | 1 | ✓ | 1 | ✓ | 1 | 1 | 1 | 1 | 1 | 1 |
| E365596 | GFPT1 | 68935 | ✓ | 44 | ✓ | 2 | ✓ | 6 | ✓ | 6 | 1 | 1 | 1 | 1 | 1 |
| E859715 | GFPT1 | 81527 | ✓ | 102 | ✓ | 12 | ✓ | 5 | ✓ | 1 | 5 | 1 | 1 | 1 | 1 |
| E656072 | GFPT1 | 70878 | ✓ | 22 | ✓ | 1 | ✓ | 1 | ✓ | 1 | 1 | 1 | 1 | 1 | 1 |
| E057609 | MUSK | 77287 | ✓ | 65 | ✓ | 3 | ✓ | 5 | ✓ | 6 | 1 | 1 | 1 | 1 | 1 |
| E124088 | MUSK | 41066 | ✓ | 25 | ✓ | 2 | ✓ | 1 | ✓ | 1 | 1 | 1 | 1 | 1 | 1 |
| E506841 | MUSK | 39153 | ✓ | 23 | ✓ | 2 | ✓ | 2 | ✓ | 1 | 4 | 1 | 1 | 2 | 1 |
| E735779 | DPAGT1 | 64019 | ✓ | 36 | ✓ | 2 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | ||
| E781411 | DPAGT1 | 42529 | ✓ | 39 | ✓ | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | ||
| E404068 | SLC25A1 | 72185 | ✓ | 40 | 1 | 2 | ✓ | 7 | 3 | 1 | 1 | 2 | 2 | ||
| E000058 | CLP1 | 49085 | ✓ | 15 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | |||
| E373377 | DHCR7 | 69905 | ✓ | 31 | 0 | ✓ | 1 | ✓ | 3 | 4 | 1 | 1 | 1 | 1 | |
| E774726 | HSPG2 | 34307 | ✓ | 53 | ✓ | 1 | ✓ | 3 | ✓ | 2 | 18 | 1 | 1 | 1 | 1 |
| E000049 | POLG | 66367 | ✓ | 17 | ✓ | 1 | ✓ | 1 | ✓ | 1 | 2 | 1 | 1 | 1 | 1 |
| E818898 | TOR1A | 66969 | ✓ | 15 | ✓ | 1 | 1 | 0 | 1 | 1 | 1 | 2 | 2 | ||
| E144500 | COL13A1 | 72939 | ✓ | 66 | 2 | ✓ | 6 | ✓ | 3 | 2 | 4 | 5 | 1 | 1 | |
| E031336 | CTNND2 | 69384 | ✓ | 32 | 4 | 2 | 1 | 1 | 2 | 1 | 2 | 2 | |||
| E050554 | TENM2 | 25626 | ✓ | 25 | 0 | 1 | 0 | 3 | 4 | 4 | 10 | 10 | |||
| Means | 34 | 2.17 | 2.24 | 2.07 | 2.31 | 1.24 | 1.28 | 1.45 | 1.41 | ||||||
| Totals | 29/29 | 23/29 | 22/29 | 23/29 | |||||||||||
- Abbreviations: HPO, Human Phenotype Ontology; NMD, nonsense-mediated decay.
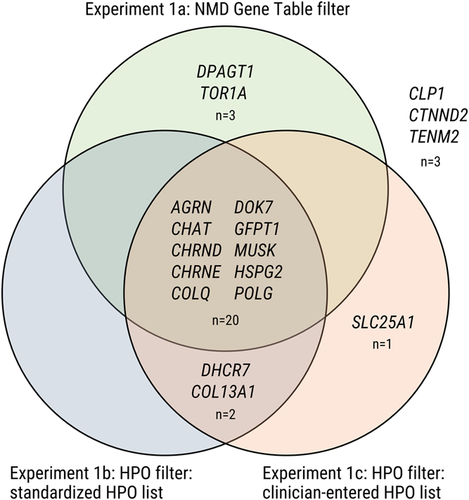
Venn diagram of gene list experiments. Experiments 1a, 1b, and 1c each returned a subset of the total number of known causative variants. Filtering with the 2016 version of the neuromuscular gene table as a virtual panel (416 genes) retained the confirmed causative gene in 23 of 29 cases, while the lists based on HPO terms retained the confirmed causative gene in 22/29 cases (standardized HPO list resulting in a list of 616 genes) and 23/29 cases (real HPO list for each case) respectively. In 3/29 cases the causative gene was not returned by any of the three gene-list methods. The diagram shows the overlap between the three methods. HPO, Human Phenotype Ontology; NMD, nonsense-mediated decay
3.1 Overall results and performance of gene list filters
As expected, before filtering, all exomes started with large numbers of homozygous variants (mean: 59,236; range: 25,626–81,527). After filtering by variant class high/moderate/low (i.e., excluding modifiers) and setting a population frequency filter of 0.001 in ExAC (Lek et al., 2016) and filtering by homozygous inheritance pattern, the variant number reduced to a more manageable size (mean: 34 variants; range: 6–102) with the causative variant still present in all cases. In Experiment 1a, filtering with the 2016 version of the neuromuscular gene table as a virtual panel (416 genes) retained the confirmed causative gene in 23 of 29 cases, resulting in a detection rate of 79.3%. Of the six cases where the neuromuscular gene table did not identify the causative gene as a possibility, in three cases it provided no candidates, while in three more cases it provided other incorrect genes while excluding the correct one (false positives). The lists based on HPO terms retained the confirmed causative gene in 22/29 cases (standardized HPO list resulting in a list of 616 genes) and 23/29 cases (real HPO list for each case), detection rates of 75.9% and 79.3%, respectively, very similar to the neuromuscular gene table results. As expected, the gene list filters were unable to pick up the new CMS genes in the cohort and this strategy thus naturally fails in a discovery paradigm. Of the cases where the correct result was not returned, the standardized HPO list provided false positives in five of seven cases (standardized list) and two of six cases (real list). The overall false positive rate is 10.3% for the neuromuscular gene table list, 17.2% for the standardized HPO list, and 6.8% for the real HPO list. In 3/29 cases the causative gene was not returned by any of the three gene-list methods.
3.2 Exomiser
The Exomiser results are not directly comparable with the gene list results, since Exomiser assigns variants a priority score rather than including or excluding them based on a predefined list. When considering the variant alone, Exomiser provides a variant score based on population frequency and pathogenicity inferences. Using this score and the associated ranking compared with the other variants evaluated, we could assess how well Exomiser performed in the absence of phenotypic data, and thus what added value the phenotypic data provided. We see (Table 3) that using variant data alone, the confirmed causative variant ranked in the first position in 18 out of 29 cases (62%). With all seven HPO terms considered and using the (human data) PhenIX algorithm, this increased to 26 out of 29 cases (89.6%). This indicates that even though we are dealing with variants that have been prefiltered to be highly plausible, the addition of phenotypic data does improve the ranking of the correct variant. Our experimental design then enabled us to look more deeply into the data to analyze the numbers and combinations of terms and genes that performed well or poorly, and the difference between human and animal data.
3.3 Exomiser with known CMS genes: Using PhenIX (Experiment 2a)
Analyzing the Exomiser rankings and scores in further detail provided some findings that allowed us to further stratify the data. Exomiser results are provided to the user with a ranking according to a combined score ranging from 0 to 1 that combines both the variant and the phenotypic score, but the variant-only score and phenotype-only score are also presented. Our results with the PhenIX algorithm showed a global difference between known CMS genes and those not associated with CMS, as is to be expected from the gene:phenotype annotations on which Exomiser bases its human-data algorithms. We therefore analyzed the known genes separately and found that although the variant-only ranks were already generally high, reflecting a strong candidate from the genotypic perspective, the ranking of the known CMS genes did further improve even with a small number of phenotypic annotations: on adding a single HPO term, the mean rank improved from 1.6 (variant alone) to 1.09 (one HPO annotation). In addition, we found that even with these high ranks, adding further HPO terms progressively improved the rank until all cases had achieved a rank of 1.0 (Figure 3a), which occurred at 6 HPO terms.
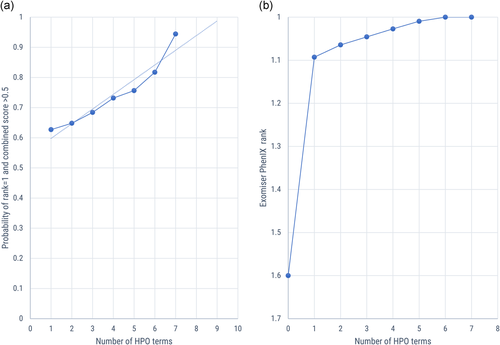
Effect of increasing the number of phenotypic annotations. (a) Change in mean rank of causative variant based on Exomiser PhenIX combined score with increasing number of HPO annotations (known CMS genes). (b) Probability of causative variant achieving a rank of 1 and score > 0.5 (known CMS genes). CMS, congenital myasthenic syndrome; HPO, Human Phenotype Ontology
However, evaluating not only the rank but also the score itself is also an important part of the analysis of the results: while the rank is a measure of the performance relative to the other variants being analyzed, the score is a measure of that variant's estimated pathogenicity and its match with the phenotype completely independently of the other variants present. Given that a “perfect” score would be 1.0 and that highly plausible variants in known disease genes frequently achieve combined scores as high as 0.997, it is evident that a low score of for example, 0.02 would raise questions about the likelihood of a variant's pathogenicity even if it were in first position in terms of ranking. Across all cases with known CMS genes, the score with a single HPO annotation ranged from 0.0695 to 0.923 (minimum and maximum mean values). It is evident from these results that it is perfectly possible for a variant to achieve a top score with even a single phenotypic annotation if this annotation happened to “hit the mark” and match the gene:phenotype annotations perfectly. It is also clear that with fewer annotations, the algorithm may be misled. We, therefore, also wanted to assess whether additional phenotypic annotation increased not only the rank but the likelihood of achieving a plausible score. Setting a value of 0.5 for the combined score as a reasonable threshold derived from user experience and combining that with the desirability of achieving a top rank, we showed that the probability of achieving both a score > 0.5 and a rank of 1 increased from 0.63 to 0.94 as the number of HPO annotations increased from one term to seven (Figure 3b). Extrapolating the trend line with higher HPO numbers suggests that the probability would reach unity at around nine HPO terms. Figure 3 further suggests that increasing the number of HPO terms to five or above increases the reliability of the analysis when it comes to known genes: not that it is not possible to achieve a high rank and score with fewer, but rather that the outcome is less likely to be incorrectly skewed by a phenotypic term that happens not to be associated with the gene in question. In addition to the progressive increase in the score of the known causative variant with increased annotation numbers, we observed a similar progressive decrease in the mean score of the second-ranked variant when analyzing the subset of cases where the correct variant was consistently ranked top. When going from one to seven annotations, the mean score of the top-ranked variant increased from 0.50 to 0.57, while that of the second-ranked variant decreased from 0.059 to 0.017 (Table S2 and Figure S1). This shows that additional annotation has the effect of increasing the confidence in the top result by increasing the distance between the top and second-ranked variants.
We then looked into the maximum and minimum scores obtained across all combinations to assess the variability of the combined score with different levels of phenotypic annotation. For each case we plotted the highest and lowest scores achieved with any number of HPO annotations from the standardized set, as well as the score achieved with the real submitter-entered HPO annotations, and the highest standardized score with a number of HPO terms equal to the number entered by the submitter. Where the number of submitter-entered terms was greater than the maximum available in the standardized list (seven terms), we used the result for seven. The results, illustrated in Figure 4, show that while the maximum achieved with any combination of terms is the best performing result (highest or equal highest in 100% of cases), the score achieved with the real HPO terms outperforms the equivalent number of standardized terms in the majority of cases (78%). This is a useful check on the use of simulated annotations, as discussed below.
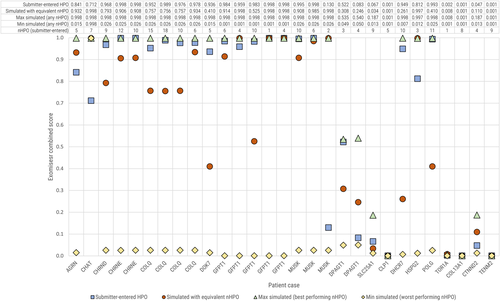
Case-by-case comparison of Exomiser PhenIX combined score values for different HPO configurations. The Exomiser combined score varied substantially depending on the HPO terms used. We show the minimum and maximum scores achieved irrespective of the number of HPO terms, as well as the score achieved with the clinician-entered HPO terms for that case, and the maximum score achieved with an equivalent number of terms from the standardized set. HPO, Human Phenotype Ontology
Repeating the analysis using the parent and grandparent terms of our originally selected terms showed that the mean Exomiser combined score increased as we moved up in the hierarchy, that is, the broader terms (lower information content) resulted in a higher score for the confirmed causative variant. The mean score across the entire cohort increased from 0.48 to 0.71 to 0.78 (original terms to parent to grandparent terms). This held true across all numbers of HPO annotations and when evaluating both the entire cohort and the subcohort comprised of known CMS genes (Table 4b). However, it was not reflected in the mean rank of the causative variant, which showed a global worsening from original to parent to grandparent across all annotation numbers. When assessed with the known gene subcohort, our original set of terms had shown a clear trend of improvement in ranking as number of annotations increased, as described above. This trend was absent in the analysis of the mean ranks with the parent terms, while with the grandparent terms the trend was reversed and showed a slight worsening in mean rank with increasing annotation levels. Evaluating the scores of the second-ranked variants in the cohort where the known causative variant was consistently ranked top showed that the combined score of the second-ranked variant also consistently increased from original to parent to grandparent term use, thus showing that the use of less specific terms increases the scores of the incorrect variant as well as the correct one.
| Number of HPO annotations | Mean rank of causative variant with PhenIX (known genes) | Mean rank of causative variant with PhenIX (new genes) | Probability of achieving rank = 1 and score > 0.5 (known genes) |
|---|---|---|---|
| 0 | 1.6 | 4.25 | N/A |
| 1 | 1.092857143 | 2.107142857 | 0.626984127 |
| 2 | 1.064285714 | 1.916666667 | 0.648148148 |
| 3 | 1.045714286 | 1.914285714 | 0.684126984 |
| 4 | 1.027142857 | 1.885714286 | 0.731746032 |
| 5 | 1.00952381 | 1.863095238 | 0.756613757 |
| 6 | 1 | 1.875 | 0.817460317 |
| 7 | 1 | 1.875 | 0.944444444 |
- Abbreviation: HPO, Human Phenotype Ontology.
| Mean score of causative variant with PhenIX (whole cohort) | Mean rank of causative variant with PhenIX (whole cohort) | Mean score of causative variant with PhenIX (known genes) | Mean rank of causative variant with PhenIX (known genes) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of HPO annotations | Original HPO terms | Parent HPO terms | Grandparent HPO terms | Original HPO terms | Parent HPO terms | Grandparent HPO terms | Original HPO terms | Parent HPO terms | Grandparent HPO terms | Original HPO terms | Parent HPO terms | Grandparent HPO terms |
| 1 | 0.23 | 0.55 | 0.81 | 1.37 | 1.38 | 1.41 | 0.60 | 0.82 | 0.92 | 1.09 | 1.09 | 1.11 |
| 2 | 0.49 | 0.78 | 0.80 | 1.30 | 1.39 | 1.47 | 0.60 | 0.82 | 0.91 | 1.06 | 1.12 | 1.18 |
| 3 | 0.35 | 0.54 | 0.61 | 1.28 | 1.39 | 1.49 | 0.62 | 0.85 | 0.89 | 1.05 | 1.13 | 1.22 |
| 4 | 0.59 | 0.77 | 0.86 | 1.26 | 1.39 | 1.52 | 0.63 | 0.87 | 0.88 | 1.03 | 1.11 | 1.26 |
| 5 | 0.56 | 0.85 | 0.90 | 1.24 | 1.38 | 1.55 | 0.65 | 0.88 | 0.88 | 1.01 | 1.10 | 1.30 |
| 6 | 0.59 | 0.77 | 0.77 | 1.24 | 1.37 | 1.56 | 0.66 | 0.90 | 0.88 | 1.00 | 1.09 | 1.31 |
| 7 | 0.72 | 0.94 | 0.97 | 1.24 | 1.38 | 1.55 | 0.68 | 0.91 | 0.89 | 1.00 | 1.10 | 1.30 |
- Abbreviation: HPO, Human Phenotype Ontology.
3.4 Genes not previously associated with CMS and inclusion of model organism and interactome comparisons (experiment 2b)
The mean rank of the variant-only score for the genes not previously associated with CMS (new CMS genes) was 4.25. When assessing these genes with the human data PhenIX filter, the rank did improve on addition of HPO terms (Table 4a), but not to the same levels as with the known genes. The improvement was due to the contribution of the variants in CTNND2, DHCR7, POLG, SLC25A1, and TOR1A, all of which do have a different human disease associated (Table 1) and some moderate overlap in phenotype with CMS. It is the cases that have not previously been associated with the human disease under examination where one might expect the alternative Exomiser algorithm, hiPHIVE (which combines human, animal and interactome data) to be of interest, since the effects of gene defects are often observed and published in animal models before a human disease analogue is found, and understanding gene function on the basis of relationships with other genes in related pathways is a common approach. We compared the Exomiser combined score obtained with the PhenIX algorithm with that obtained from the hiPHIVE algorithm and stratified the results by known versus new CMS gene. Our results show that mean scores from the known CMS genes increased from 0.627 to 0.944 when moving from PhenIX to hiPHIVE, while those from the non-CMS genes increased from 0.161 to 0.688. All cases showed this increase individually, and cases that scored in the lower range with PhenIX tended to increase more substantially with hiPHIVE, resulting in a convergence of scores at the higher range (Figure 5). This indicates that including animal and interactome data does increase the level of evidence about a particular gene:phenotype correlation for both known and novel genes. However, an important caveat is that we did not observe a corresponding improvement in rank of the known causative variant, because the score of other variants in the analysis was also increased with the addition of the animal and interactome data. Across all experiments and all variants included in the ranking (i.e., every variant considered and not purely the causative variant), the mean Exomiser combined score was 0.0389 with PhenIX and 0.334 with hiPHIVE—a tenfold increase. Thus we see that the Exomiser combined score increases across the board when moving from the PhenIX to the hiPHIVE algorithm, so it is important not to draw conclusions purely from the increase.
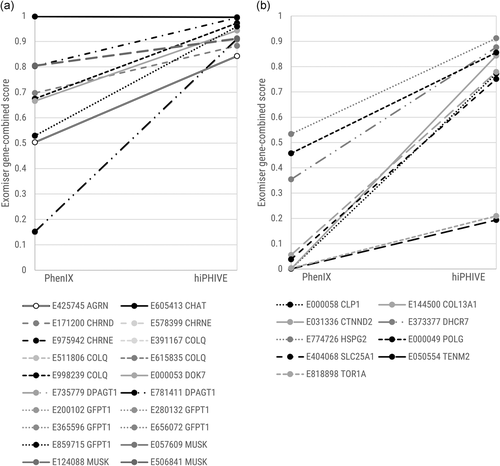
Comparison of combined score between PhenIX and hiPHIVE algorithms. (a) Change in combined score when changing algorithm from PhenIX to hiPHIVE: known CMS genes. (b) Change in combined score when changing algorithm from PhenIX to hiPHIVE: genes not previously associated with CMS. CMS, congenital myasthenic syndrome
To find the reasons for the cases in which the algorithms did not rank our known gene in first position, we need to look at the individual cases and examine not only the score of the causative gene but also the other variants present. Looking at the individual genes, we see that COL13A1 is one where the addition of the animal and interactome data improved the performance, as the rank improved from 4 (PhenIX) to 1 (hiPHIVE). This gene is in fact a recent CMS gene, (Logan et al., 2015) now listed in OMIM and with new cases just published (Dusl et al., 2019). However, when analyzed against human data alone the Exomiser gene:pheno score was zero, thus it is evidently too recent to have been included in the gene:phenotype correlation used by Exomiser at the time of running the analysis, while the data from animal models and interactome data apparently makes the variant much more convincing, resulting in a score increase from 0.000849 to 0.843. In contrast, the rank of TENM2 worsened from 4 to 10, despite its absolute score increasing from 0.000812 to 0.194. The top scoring gene in this case, RFC3, received a score of 0.757. Clearly, then, even animal model and interactome data does not provide a high level of evidence for this gene as a cause of the phenotype. The third gene to show a change, SLC25A1, was ranked first by the PhenIX algorithm but second by the hiPHIVE algorithm. With hiPHIVE, the scores of the top three variants cluster closely together at 0.749, 0.747, and 0.746, indicating that the algorithm has very similar levels of evidence for each. Like COL13A1, SLC25A1 is a relatively recent CMS gene (Chaouch et al., 2014) and the evidence in databases is lacking. However, when annotations are added, Exomiser's performance does improve correspondingly. We ascertained that COL13A1 annotations had been added to the HPO gene:phenotype annotations after we had performed our initial experiments and therefore chose to re-run the experiment for COL13A1 with seven HPO terms and the PhenIX algorithm using the February, 2019 annotation set. This resulted in a highly convincing rank of 1 and combined score of 0.998 for our known causative variant. This chance opportunity to test the effect of the addition of highly relevant annotations confirms the ability of the Exomiser algorithm to respond when the relevant data is available to it.
4 DISCUSSION
From a practical user perspective, we are interested in how to maximize the likelihood that a user will be able to correctly identify the causative variant, and to minimize the time this takes. Our variant class and population frequency filters dramatically reduced the number of variants under consideration, and we consider this to be a necessary first step before considering phenotype. Our intentional selection of homozygous recessive cases, which was done to restrict the genotypic variables in our experiment set, was an added constraint on variant numbers: we would expect larger numbers of variants to remain after the variant-based filtering if we had chosen cases with a dominant or compound heterozygous inheritance pattern. Additional variant-based filters such as those based on pathogenicity inferences are often also applied either before or after phenotype-based filtering, but we did not use these to observe the tool performance in the absence of this restriction.
Once we arrive at consideration of the phenotype, our results confirm that as a first quick phenotype-based filter, using a virtual panel like the neuromuscular gene table is an effective strategy to rapidly determine whether there are candidate variants in a gene already associated with a related disease. This is an approach familiar to many clinicians from a diagnostics-based workflow and panel sequencing, where a diagnosis will typically only be returned if it is in a confirmed disease gene. While we cannot extrapolate from our highly constrained CMS cohort to other rare diseases, it is noteworthy that in our case the HPO-based gene list provides comparable detection rates to the curated gene list. This is an important finding because not all disease areas have a comparable curated and regularly updated gene list available, and in these cases the option of generating a customized HPO-based phenotypic filter may be particularly valuable. Other studies with diagnostic exomes in a wider range of phenotypes have found HPO-based filtering to have high detection rates for known genes, with one study on 55 cases finding this strategy included the correct variant among the returned results in 100% of cases (Kernohan et al., 2018).
It is worth mentioning here that curated disease-gene lists like the neuromuscular gene table tend to be updated only periodically and thus may often lag behind the latest discoveries by months or years, and this can affect results: two of our six negative results (COL13A1 and SLC25A1), where the confirmed causative variant was not returned, would have become positive if the very latest version of the neuromuscular gene table had been used, but this is not yet available within the RD-Connect system owing to the recency of the update (December, 2018; Bonne, Rivier, & Hamroun, 2018). Despite being generated on the fly from the phenotypic input, the HPO-based list is not immune to this effect, and also returned negative results in the case of some more recent known disease genes. This may be explained by the fact that the correlation between the phenotype and the gene still relies on curated gene:phenotype annotations in online databases, and while the annotations produced by the HPO developers are updated monthly, they nevertheless rely on extraction of data from published literature and online genetics databases and a delay is to be expected. Disease genes with atypical phenotypes are also less likely to be captured by either method, as they will not have been included in the curation effort for a domain-specific list and may not have been annotated with the relevant phenotype for gene:phenotype correlation.
It is important to note that in cases where more than one result is returned, neither the disease-gene list nor the HPO-based list method makes any attempt to rank or otherwise quantify the variants returned by likelihood of causality; they merely express whether a variant has been found in a gene associated with the disease phenotype or not. The user must then assess likelihood by following up each candidate by other methods such as pathogenicity prediction using in silico tools, segregation analysis where family members are available, and Sanger sequencing confirmation. Within our use case of the RD-Connect interface, segregation analysis is possible for all family members whose data is in the system, and the results of pathogenicity prediction tools such as PolyPhen2, MutationTaster, and SIFT and the CADD score for the variant are presented to the user alongside the variant results, making this further refinement a fairly rapid process, but this may be a more manual process where an analysis system of this kind is not in use.
While over half of cases returned more than one candidate for further investigation, we did not consider this an inaccurate result in itself, since it is standard practice to follow up multiple candidates. However, the cases in which candidates were returned but the correct candidate was not among them, which we have termed false positives in the results above, are worthy of note when using a gene list filter, since it is more misleading for the user if incorrect results are returned than if nothing is returned—the latter clearly shows that further investigation of other genes is necessary, while false positives may lead the user further down the wrong track.
Exomiser is a more flexible tool than a simple gene-list filter. By combining an assessment of the variant pathogenicity with an algorithm for weighting based on phenotypic matching, it provides the user with more specific data on which to base further investigations. When using human data alone, it performs very well with known genes, ranking the known causative variant top in 100% of cases when run with six or more HPO terms. It is unavoidably still affected by the same biases that affect the gene-list filters, in that a gene without previous human disease association cannot achieve a high phenotypic score. However, it is less likely to exclude the causative variant completely even when it does not rank it highly. Its results may be skewed with smaller numbers of phenotypic annotations, as we see in the variation in combined score between 0.007 and 0.922 (average minimum-maximum across all known genes when run with a single phenotypic annotation). This variation presumably occurs because the algorithm returns a high result when the single annotation happens to perfectly match a database annotation and a low result when the annotation is not present. Adding more phenotypic annotations decreases the likelihood of any single mismatch throwing the algorithm off.
Exomiser's algorithm is particularly helpful in less straightforward cases, with newer genes or those with an unusual phenotype. Of the nine cases where the correct variant was excluded by at least one of the gene list filters, Exomiser with its phenotypic rating nevertheless ranked that variant top in six cases, and if we extend this to both PhenIX and hiPHIVE and look at both top and second-ranked variants, then Exomiser met the criteria in eight cases out of nine. Given that in a standard workflow the user would be likely to follow up at least the top three cases at minimum, this is a strong result for cases with less common diagnostic variants. In our study, all the cases with known genes (both the typical CMS genes as well as the other disease genes with overlapping phenotypes) were confirmed by the treating clinician including segregation and full phenotype review. This is also true of the newer CMS genes, COL13A1 and SLC25A1. The genes that have not been related to human disease remain candidate genes until a second, confirmatory family with the same genetic defect has been identified and functional assays have been carried out. Here, too, phenotypic approaches are also increasingly used in algorithms to assess similarity between patients (Haendel, Chute, & Robinson, 2018) and “matchmaker” APIs attempt to use this method to find confirmatory cases across multiple databases (Buske, Schiettecatte et al., 2015).
From the end-user perspective it is helpful to know how many phenotypic annotations are required for a reliable outcome. Varying the number of HPO terms from the standardized list in the Exomiser analysis enabled us to show that even a single HPO annotation improved the mean ranking of the variant over that obtained using genotypic data alone, but that the algorithm was susceptible to being skewed by insufficient data. In our cohort, annotation with six or more terms ranked the known causative variant top in all cases when dealing with known genes, and with seven terms the probability of achieving both a rank of 1 and a combined score above 0.5 was 0.94, thus suggesting that while it is desirable to annotate with more than five terms, going above nine may not add much value, at least in our disease area. However, since we also showed that repeating the analysis with less specific annotation sets slightly decreased the mean rank obtained and reversed the trend of improving rank with increasing annotation even while the mean combined score increased, it is evident that the choice of specificity of annotation does make a difference, which is important for the clinical annotator to know from a practical perspective. Our original terms were chosen without explicit reference to their IC but rather as a real example of terms that CMS experts would frequently use in clinical practice, and from this limited experiment it would appear that this may achieve better ranking results than requesting annotators to intentionally select broader terms. Our analysis of the second-ranked variants showed that the use of less specific annotation increased the scores of incorrect variants as well as the known causative variant, presumably because less specific annotation acts as more of a general “catch-all.” This further demonstrates that the absolute score must be interpreted with caution and always viewed in the context of the scores and ranks of the other variants. These points would need further evaluation in a larger cohort to make confident recommendations, but from our limited data set it would appear that greater specificity in annotation is preferable and that highly unspecific terms may even lead to poorer performance. It should be noted that the algorithm still ranked the known causative variant top in the majority of cases even with the use of less specific terms, which is indicative of its robustness.
Our study intentionally restricted the cohort evaluated to be able to apply a standard set of phenotypic annotations across all cases and to reduce the number of genotypic variables. This limits its applicability to other disease groups and other inheritance patterns. Further research to extend the concept will be of value and may consist both in repeating the phenotypic variation tests in a similarly limited cohort in a different disease area and in assessing a larger cohort of unsolved cases without limitation to a disease area using only the clinician-entered phenotypic annotations and not a controlled set as in our study. Further, while our study did intentionally include a small number of novel CMS genes to enable a comparison between a diagnostic paradigm with known genes and a discovery paradigm with novel genes, this did not make up a large number of cases in our study, and only two had no prior human disease association at all. It would thus be valuable to perform a trial of Exomiser with its hiPHIVE algorithm on a larger cohort of cases solved for the first time with a novel gene. This may become possible through the case aggregation taking place in RD-Connect through projects like BBMRI-LPC, Solve-RD (https://solve-rd.eu) and the European Joint Programme for Rare Disease (http://www.ejprarediseases.org/), and internationally in Care4Rare Solve (Boycott et al., 2017). These projects also aim to solve some of the most challenging unsolved rare disease cases, and good phenotypic annotation is expected to play an important role. It is likely to be useful for conditions with suspected dominant or sporadic inheritance, which usually have a large number of rare heterozygous variants that need to be compared and prioritized. Simulating the optimal number of HPO terms for these and other more complex inheritance patterns is warranted but was beyond the scope of this study.
In the discovery paradigm, there are of course other in silico strategies for variant prioritization open to the researcher, including assessment of tissue expression and protein:protein interactions, other tools incorporating phenotype (Pengelly et al., 2017), and strategies for uncovering implicit associations between gene and functional effects even when these have not yet been published through direct evidence (Hettne et al., 2016). The tools studied here may only form a part of the analysis strategy in such cases, and functional analysis in model organisms as well as confirmatory cases from second families remains the mainstay of confirming a novel gene discovery. However, we have shown that phenotype is a highly valuable prioritization strategy in the diagnostic paradigm, and that it also adds value in the discovery paradigm when Exomiser is used. These tools depend on annotation of gene:phenotype relationships in databases derived from associations curated from the published literature. New methods for extracting and packaging these relationships such as those offered by phenopackets (Mungall, 2016) will improve the performance of the tools by increasing the data they use as input, particularly when combined with the rapid growth of databases like RD-Connect where genomic data is associated with HPO-coded phenotypic profiles. Both in the discovery paradigm and in cases with nonspecific or unusual phenotypic presentation, the use of Exomiser provides added value over a simple gene list filter, since it provides a more sophisticated prioritization method rather than simple inclusion or exclusion. In all cases, the benefit of the tools is the way they enable the user to filter out extraneous information and quickly approach a small subset of strong candidates for follow-up, rather than replacing the human user in the diagnostic process. Overall, our results therefore still speak to the necessity for expert evaluation in all cases, both at the initial patient encounter when phenotyping and coding is performed as well as during the interpretation and re-evaluation of sequencing data in the context of phenotype.
ACKNOWLEDGEMENTS
Data was analyzed using the RD-Connect Genome-Phenome Analysis Platform developed under the 2012-2018 FP7-funded project RD-Connect (Grant Agreement No.: 305444). R. T., S. B., A. P. N., P. tH., and H. L. received support from the EU FP7 projects 305444 (RD-Connect) and 305121 (NeurOmics). S. B. received support from the EU FP7 project 313010 (BBMRI-LPC). S. B. and P. tH. received support from the EU Horizon 2020 project 779257 (Solve-RD). S. B., P. tH., and H. L. received support from the EU Horizon 2020 project 825575 (EJP-RD). H. L. is supported by a project grant of the Canadian Institutes of Health Research (CIHR PJT 162265). We gratefully acknowledge Steve Laurie and Davide Piscia (CNAG-CRG) for exome data processing and support with the RD-Connect GPAP, including the implementation of functionality that has been key for the execution of the study. We are extremely grateful to the patients and families who consented to the use of their data for research and the clinical submitters of the cases analyzed, including Atchayaram Nalini, Veeramani Preethish-Kumar, Seena Vengalil, and Saraswati Nashi (Bangalore, India).